|
|
| (29 intermediate revisions by 2 users not shown) |
| Line 2: |
Line 2: |
| | | | |
| | In this chapter. . .<br /> | | In this chapter. . .<br /> |
| − | [["Supported graphic formats overview"]]<br />
| + | *Supported graphic formats overview |
| − | [["Understanding the MIPAV dimensionality and coordinate system" ]] <br />
| + | *Understanding the MIPAV dimensionality and coordinate system |
| − | [["Data types"]]<br />
| + | *Data types |
| − | [["Headers" ]]<br />
| + | *Headers |
| − | [["Endianness"]]<br />
| + | *Endianness |
| − | [["Viewing and editing image attributes" ]]<br />
| + | *Viewing and editing image attributes |
| | | | |
| | This chapter also explains the dimensionality and coordinate system used by MIPAV, the data types MIPAV supports, the header offset information, endianness, and how to view and edit image attributes.It also provides a basic information about image file formats supported by MIPAV. More information about graphic file formats and the formats that MIPAV supports can be found in Appendix C: "Supported formats" . | | This chapter also explains the dimensionality and coordinate system used by MIPAV, the data types MIPAV supports, the header offset information, endianness, and how to view and edit image attributes.It also provides a basic information about image file formats supported by MIPAV. More information about graphic file formats and the formats that MIPAV supports can be found in Appendix C: "Supported formats" . |
| | | | |
| | == Supported graphic formats overview == | | == Supported graphic formats overview == |
| − | | + | MIPAV supports three categories of graphic file formats: vector (i.e., volume of interest, or VOI), bitmap (i.e., 2D to 5D grayscale or RGB images), and 3D images.<br />'''Vector file formats ''' are usually used to store line-based elements. These elements can be geometric shapes, such as polygons, curves, or splines. MIPAV can read and generate several vector-formatted file types including the MIPAV lookup table (LUT), MIPAV MTX, MIPAV Graphics Plot (PLT), and MIPAV VOI. See also [[Supported Formats|Appendix C: "Supported formats" ]].<br />'''Bitmap file formats''' are usually used to store real-world graphics data e.g., photographs or medical images such as CT scans. The data portion of bitmap files contains numerical data that indicates the color of each pixel or voxel in the image. MIPAV can read and generate a number of bitmap formatted file types, such as Adobe Photoshop (PSD), Graphics Interchange File (GIF), Sun Raster (RS), Tagged Image File Format (TIFF), and Truevision Graphics Adapter (TGA).<br /></font>'''MIPAV uses 3D graphic formatted files''' to store descriptions of the color and shape of 3D models of real-world and imaginary objects.<br /> |
| − | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> MIPAV supports three categories of graphic file formats: vector (i.e., volume of interest, or VOI), bitmap (i.e., 2D to 5D grayscale or RGB images), and 3D images.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline">'''<font color="#000000">Vector file formats </font>'''</span>are usually used to store line-based elements. These elements can be geometric shapes, such as polygons, curves, or splines. MIPAV can read and generate several vector-formatted file types including the MIPAV lookup table (LUT), MIPAV MTX, MIPAV Graphics Plot (PLT), and MIPAV VOI. See also [[Appendix C: "Supported formats" ]].<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline">'''<font color="#000000">Bitmap file formats</font>'''</span> are usually used to store real-world graphics data e.g., photographs or medical images such as CT scans. The data portion of bitmap files contains numerical data that indicates the color of each pixel or voxel in the image. MIPAV can read and generate a number of bitmap formatted file types, such as Adobe Photoshop (PSD), Graphics Interchange File (GIF), Sun Raster (RS), Tagged Image File Format (TIFF), and Truevision Graphics Adapter (TGA).<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline">'''<font color="#000000">MIPAV uses 3D graphic formatted files </font>'''</span>to store descriptions of the color and shape of 3D models of real-world and imaginary objects.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> To support the wide range of image-processing needs of the NIH intramural research community, MIPAV reads and writes the image file types indicated in Table 1, refer to [[Appendix C: "Supported formats"]]. <br /></font></div>
| + | To support the wide range of image-processing needs of the NIH intramural research community, MIPAV reads and writes the image file types indicated in Table 1, refer to [[Supported Formats |Appendix C: "Supported formats"]]. <br /> |
| | | | |
| | == Understanding the MIPAV dimensionality and coordinate system == | | == Understanding the MIPAV dimensionality and coordinate system == |
| | + | Internally, MIPAV is designed to store images of any dimensionality and thus is ''n'' dimensional. Figure 1 is a model of an example 3D-image where each position within the grid represents a ''voxel''.<br />'''voxel'''-The smallest distinguishable cube-shaped part of a 3D-image. <br /> |
| | | | |
| − | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> Internally, MIPAV is designed to store images of any dimensionality and thus is <span style="font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">''<font color="#000000">n </font>''</span>dimensional. Figure 1 is a model of an example 3D-image where each position within the grid represents a <span style="font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">''<font color="#000000">voxel</font>''</span>.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 5pt; margin-left: 0pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline">'''<font color="#000000">voxel</font>'''</span>-The smallest distinguishable cube-shaped part of a 3D-image. <br /></font></div>
| + | The voxel in the upper left-most position is considered the origin of the dataset. (A volumetric dataset is a set of cross-sectional images). Ideally, each voxel would be isotropic (i.e., cubic), but clinical datasets are often acquired in such a manner that the resolution in the ''x'' and ''y'' dimensions is greater than the resolution in the <span style="font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">''<font color="#000000">z</font>''</span> dimension, thus producing rectangular voxels (elongated in the ''z''. This voxel anisotropy is taken into account in the execution of the algorithms found in MIPAV. |
| | | | |
| − | The voxel in the upper left-most position is considered the origin of the dataset. (A volumetric dataset is a set of cross-sectional images). Ideally, each voxel would be isotropic (i.e., cubic), but clinical datasets are often acquired in such a manner that the resolution in the <span style="font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">''<font color="#000000">x</font>''</span> and <span style="font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">''<font color="#000000">y</font>''</span> dimensions is greater than the resolution in the <span style="font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">''<font color="#000000">z</font>''</span> dimension, thus producing rectangular voxels (elongated in the <span style="font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">''<font color="#000000">z</font>''</span> direction). This voxel anisotropy is taken into account in the execution of the algorithms found in MIPAV.
| + | Because MIPAV's image storage buffer is ''n''-dimensional, it can store 2D, 3D, and 4D medical images. CT and MRI are examples of 3D medical images where the three dimensions (''x, y,'' and ''z'') are spatial dimensions. Other 3D datasets include fluoroscopy volumes, where the first two dimensions are spatial and the third dimension is time. |
| | | | |
| − | Because MIPAV's image storage buffer is <span style="font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">''<font color="#000000">n</font>''</span>-dimensional, it can store 2D, 3D, and 4D medical images. CT and MRI are examples of 3D medical images where the three dimensions (<span style="font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">''<font color="#000000">x, y,</font>''</span> and <span style="font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">''<font color="#000000">z</font>''</span>) are spatial dimensions. Other 3D datasets include fluoroscopy volumes, where the first two dimensions are spatial and the third dimension is time.
| |
| − |
| |
| − | <div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000">
| |
| | | | |
| | {| border="1" cellpadding="5" | | {| border="1" cellpadding="5" |
| Line 32: |
Line 30: |
| | |} | | |} |
| | | | |
| − | <br /></font></div> | + | <br /> |
| | + | |
| | + | == Re-ordering data from image to voxel order == |
| | + | |
| | + | When recorded in scanner or image order, multi-component images are stored as consecutive volumes. This is convenient for visualization, since one can easily render a particular 3D volume, however, it is inconvenient for parallel processing, as one must read the entire 4D image in order to get the specific component(s) for a particular processing. |
| | + | |
| | + | In '''voxel order''' all components for a particular voxel are stored together. This allows to read the image one voxel at a time, or skip ahead to a particular voxel, without storing the entire image into memory. |
| | + | |
| | + | See also: [[http://www.grahamwideman.com/gw/brain/orientation/orientterms.htm Orientation and Voxel-Order Terminology: RAS, LAS, LPI, RPI, XYZ and All That]]. |
| | | | |
| | == Data types == | | == Data types == |
| | | | |
| − | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> A <span style="font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">''<font color="#000000">data type</font>''</span> is the classification of a particular type of information. For example, a floating point data type indicates a number with a decimal point. Data read from image files are stored in MIPAV's data structures. MIPAV supports all the basic data types shown in Table 1 and may in the future support more composite data types including color and complex numbers. The image data read from the file is stored in a data buffer of the same basic data type, preventing the reduction of image information that results from conversion into a fixed data type.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000">
| + | A ''data type'' is the classification of a particular type of information. For example, a floating point data type indicates a number with a decimal point. Data read from image files are stored in MIPAV's data structures. MIPAV supports all the basic data types shown in Table 1 and may in the future support more composite data types including color and complex numbers. The image data read from the file is stored in a data buffer of the same basic data type, preventing the reduction of image information that results from conversion into a fixed data type.<br /> |
| | | | |
| | {| border="1" cellpadding="5" | | {| border="1" cellpadding="5" |
| Line 97: |
Line 103: |
| | |} | | |} |
| | | | |
| − | <br /></font></div> | + | <br /> |
| | | | |
| | == Headers == | | == Headers == |
| | | | |
| − | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> If you attempt to open a raw image, you may need to supply MIPAV with the <span style="font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">''<font color="#000000">header offset.</font>''</span> The header offset indicates the size of the space reserved at the beginning of the file where specific types of information is kept. This space, which is called the header, precedes the image data. If you know the length of the header, enter it in the header offset text box. When MIPAV accesses the file, it skips the header offset and begins to read the image data. <span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline">'''<font color="#000000">Note that</font>'''</span> not all image file formats have a header.<br /></font></div>
| + | If you attempt to open a raw image, you may need to supply MIPAV with the ''header offset.'' The header offset indicates the size of the space reserved at the beginning of the file where specific types of information is kept. This space, which is called the header, precedes the image data. If you know the length of the header, enter it in the header offset text box. When MIPAV accesses the file, it skips the header offset and begins to read the image data. '''Note''' that not all image file formats have a header.<br /> |
| | | | |
| | == Endianness == | | == Endianness == |
| | | | |
| − | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <span style="font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">''<font color="#000000">Endianness</font>''</span> refers to the byte ordering of the data. Some computers order the data with the least significant byte (LSB) first followed by the most significant byte (MSB). This byte order is referred as "little endian" or Intel byte ordering. machines that use little-endian byte ordering are VAXes, Intel x86, and Pentium. The reverse is MSB then LSB, which is referred as "big endian" or Motorola byte ordering. Machines that use big-endian byte ordering are IBM System 3D, RISC, and a Motorola 680x0. MIPAV is biendian; that is, it supports both big- and little-endian byte-ordering formats.<br /></font></div>
| + | ''Endianness'' refers to the byte ordering of the data. Some computers order the data with the least significant byte (LSB) first followed by the most significant byte (MSB). This byte order is referred as "little endian" or Intel byte ordering. machines that use little-endian byte ordering are VAXes, Intel x86, and Pentium. The reverse is MSB then LSB, which is referred as "big endian" or Motorola byte ordering. Machines that use big-endian byte ordering are IBM System 3D, RISC, and a Motorola 680x0. MIPAV is biendian; that is, it supports both big- and little-endian byte-ordering formats.<br /> |
| | | | |
| | == Viewing and editing image attributes == | | == Viewing and editing image attributes == |
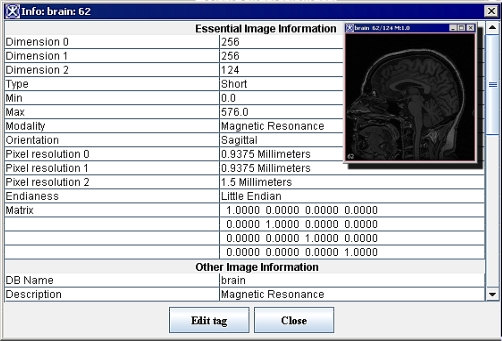
| − | | + | Information about the image appears in the Image Information dialog box and in the Image Attributes dialog box. <br /> |
| − | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> Information about the image appears in the Image Information dialog box and in the Image Attributes dialog box. <br /></font></div>
| + | |
| | | | |
| | The Image Attributes window displays the name of the image file and information about the resolution of the image. The About Image window displays more detailed information about the image, such as the orientation of the image or the number of bits per pixel or voxel. | | The Image Attributes window displays the name of the image file and information about the resolution of the image. The About Image window displays more detailed information about the image, such as the orientation of the image or the number of bits per pixel or voxel. |
| | | | |
| | === Viewing image attributes === | | === Viewing image attributes === |
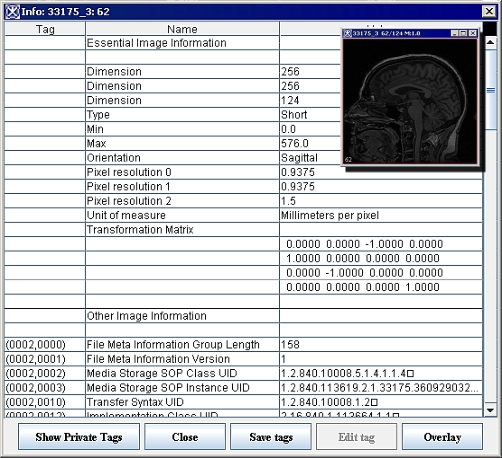
| − | | + | To view the attributes of an image, select Image > Attributes > View Header in the MIPAV window. For DICOM datasets, the Image Information dialog box similar to the one shown in Figure 2-a appears. For non-DICOM datasets, an Image Information dialog box similar to the one in Figure 2-b appears. <br /> |
| − | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> To view the attributes of an image, select Image > Attributes > View Header in the MIPAV window. For DICOM datasets, the Image Information dialog box similar to the one shown in Figure 2-a appears. For non-DICOM datasets, an Image Information dialog box similar to the one in Figure 2-b appears. <br /></font></div>
| + | |
| − | | + | |
| − | <div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000">
| + | |
| | | | |
| | {| border="1" cellpadding="5" | | {| border="1" cellpadding="5" |
| Line 136: |
Line 138: |
| | | | |
| | | | |
| − | <br /></font></div> | + | <br /> |
| | + | |
| | + | |
| | | | |
| | === Editing image attributes === | | === Editing image attributes === |
| | | | |
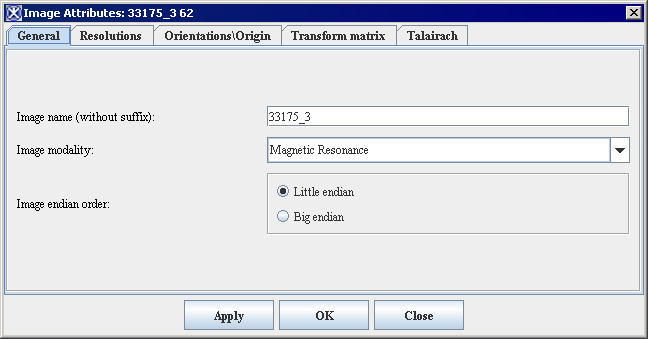
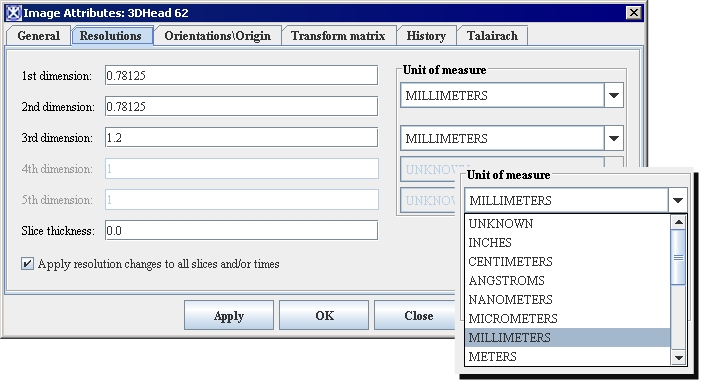
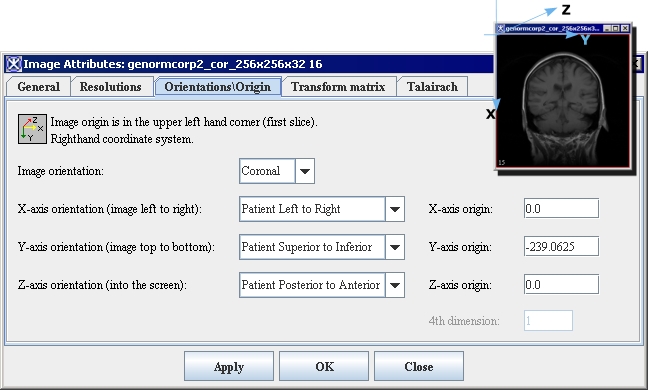
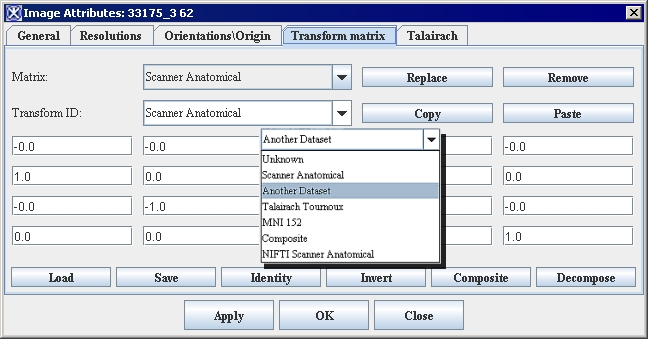
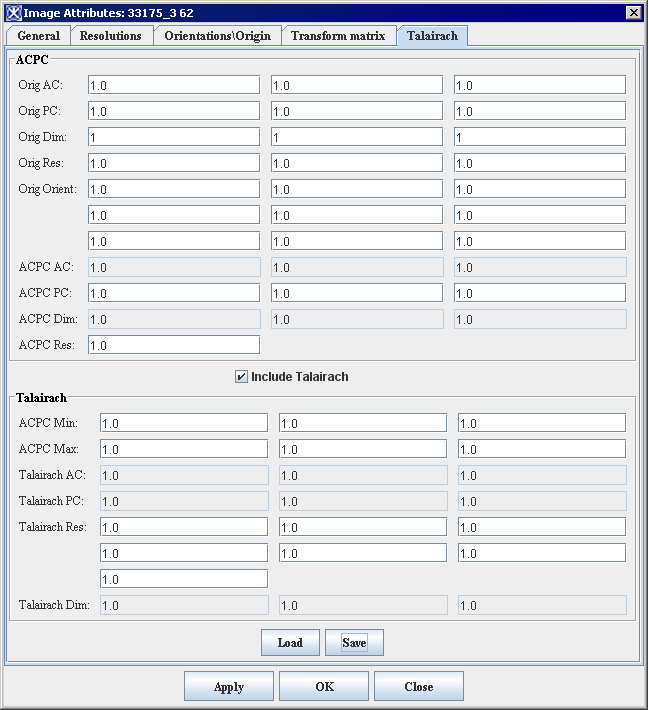
| − | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> You can edit some of the image attributes for both DICOM datasets and non-DICOM datasets in the Image Attributes dialog box (Figure 3). The Image Attributes dialog box includes five tabbed pages, which are the following:<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline">'''<font color="#000000">General page</font>'''</span> displays the name of the image (without its extension, or suffix) and lists the image modality<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline">'''<font color="#000000">Resolutions page</font>'''</span> indicates the unit of measure for each dimension<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline">'''<font color="#000000">Orientations/Origin page</font>'''</span> lists the image orientations <br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline">'''<font color="#000000">Transform Matrix</font>'''</span> displays and lists the values for the transform matrix<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline">'''<font color="#000000">Talairach</font>'''</span> displays Talairach transform <br /></font></div>
| + | You can edit some of the image attributes for both DICOM datasets and non-DICOM datasets in the Image Attributes dialog box (Figure 3). The Image Attributes dialog box includes five tabbed pages, which are the following:<br /><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline">'''<font color="#000000">General page</font>'''</span> displays the name of the image (without its extension, or suffix) and lists the image modality<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline">'''<font color="#000000">Resolutions page</font>'''</span> indicates the unit of measure for each dimension<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline">'''<font color="#000000">Orientations/Origin page</font>'''</span> lists the image orientations <br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline">'''<font color="#000000">Transform Matrix</font>'''</span> displays and lists the values for the transform matrix<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline">'''<font color="#000000">Talairach</font>'''</span> displays Talairach transform <br /></font></div> |
| | | | |
| | By default, the General page always appears first when the Image Attributes dialog box opens. | | By default, the General page always appears first when the Image Attributes dialog box opens. |
| | | | |
| | ==== Image Attributes dialog box ==== | | ==== Image Attributes dialog box ==== |
| − |
| |
| − | <div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000">
| |
| − | <div style="font-style: normal; margin-bottom: 3pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline">'''<font color="#000000"> Figure 3. The Image Attributes dialog box options <br /></font>'''</div>
| |
| − |
| |
| − |
| |
| − | <br /></font></div><div style="font-style: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 14pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline">'''<font color="#000000"> To modify the name of the image dataset<br /></font>'''</div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 1 Open either a DICOM or non-DICOM image dataset.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 2 Select Image > Attributes > Edit Attributes in the MIPAV window. The Image Attributes dialog box opens.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 3 Select the name of the image in the Image name (without suffix) box.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 4 Type a new name of the image. <br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 5 Click either Apply or OK. The new image name appears in the header on the image window.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000">
| |
| − |
| |
| − | {| border="1" cellpadding="5"
| |
| − | |+
| |
| − | |-
| |
| − | |
| |
| − | <div style="font-style: normal; margin-bottom: 1pt; margin-left: 0pt; margin-right: 0pt; margin-top: 17pt; text-align: center; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline">'''<font color="#000000"> What's the Difference Between<br />Clicking Apply or Clicking OK?<br /></font>'''</div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 7pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> Click Apply when you want the dialog box or window to remain open after it makes the change so that you can make further modifications. If instead you click OK, the dialog box or window closes immediately after making the current change.<br /></font></div>
| |
| − | |}
| |
| − |
| |
| − | <br /></font></div><div style="font-style: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 14pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline">'''<font color="#000000"> To edit the dimensions and units of measure<br /></font>'''</div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 1 Open either a DICOM or non-DICOM image dataset.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 2 Select Image > Attributes > Edit Attributes in the MIPAV window. The Image Attributes dialog box opens.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 3 Click Resolutions. The Resolutions page in the Image Attributes dialog box appears.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 4 Type the changed values in the dimension text boxes on the left and then select the appropriate value in the Unit of measure boxes on the right.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 5 Click either Apply or OK.<br />
| |
| − |
| |
| − | <div style="font-style: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 14pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline">'''<font color="#000000"> To edit the <span style="text-decoration: none; text-transform: none; vertical-align: baseline">'''''<font color="#000000">x, y,</font>'''''</span> and <span style="text-decoration: none; text-transform: none; vertical-align: baseline">'''''<font color="#000000">z </font>'''''</span>orientations of image datasets<br /></font>'''</div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 1 Open either a DICOM or non-DICOM image dataset.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 2 Select Image > Attributes > Edit Attributes in the MIPAV window. The Image Attributes dialog box opens.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 3 Click Orientations. The Orientations page (Figure 4) in the Image Attributes dialog box opens.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 4 Select the appropriate values in the boxes on the right.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 5 Click Apply or OK.<br /></font></div><div style="font-style: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 14pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline">'''<font color="#000000"> To change the origin for the first image slice <br /></font>'''</div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 1 Open either a DICOM or non-DICOM image dataset.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 2 Select Image > Attributes > Edit Attributes in the MIPAV window. The Image Attributes dialog box opens.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 3 Click Orientation. The Orientation page (Figure 4]) in the Image Attributes dialog box appears.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 4 Type the appropriate values in the dimension boxes.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 5 Click Apply or OK.<br /></font></div><div style="font-style: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 14pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline">'''<font color="#000000"> To change the transform matrix <br /></font>'''</div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 1 Open either a DICOM or non-DICOM image dataset.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 2 Select Image > Attributes > Edit Attributes in the MIPAV window. The Image Attributes dialog box opens.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 3 Click Transform Matrix. The Transform Matrix page (Figure 3) in the Image Attributes dialog box appears.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 4 Select the appropriate transform ID in the Transform ID box.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 5 Load and save transform matrices.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><font color="#000000"> 6 Click Apply or OK.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000">
| |
| − |
| |
| − | ==testing section==
| |
| | {| border="1" cellpadding="5" cellspacing="0" | | {| border="1" cellpadding="5" cellspacing="0" |
| | |+'''Figure 3. Image Attributes dialog box''' | | |+'''Figure 3. Image Attributes dialog box''' |
| | |- | | |- |
| − | | '''Apply''' | + | |'''Apply''' |
| − | | + | |
| | | rowspan="1" colspan="2" |applies the changes to the current image, but doesn't close the dialog box. | | | rowspan="1" colspan="2" |applies the changes to the current image, but doesn't close the dialog box. |
| | |- | | |- |
| | |'''OK''' | | |'''OK''' |
| − |
| |
| | | rowspan="1" colspan="2" |applies the changes to the current image and closes the dialog box. | | | rowspan="1" colspan="2" |applies the changes to the current image and closes the dialog box. |
| | |- | | |- |
| Line 180: |
Line 163: |
| | |'''Help''' | | |'''Help''' |
| | | rowspan="1" colspan="2" |opens the on-line help. | | | rowspan="1" colspan="2" |opens the on-line help. |
| − |
| |
| | |- | | |- |
| | | rowspan="1" colspan="3" |'''General tab''' | | | rowspan="1" colspan="3" |'''General tab''' |
| − |
| |
| | |- | | |- |
| − | | [[Image:ImageAttributesGeneral.jpg]] | + | | rowspan="1" colspan="3"|[[Image:ImageAttributesGeneral.jpg]] |
| | |- | | |- |
| | |Image name (without suffix) | | |Image name (without suffix) |
| Line 194: |
Line 175: |
| | |- | | |- |
| | |The list includes: | | |The list includes: |
| − |
| |
| | | rowspan="1" colspan="2" |Unknown Modality, Biomagnetic Imaging, Color Flow Doppler, Computed Radiography, Computed Tomography, Diaphanography, Digital Radiography, Duplex Doppler, Endoscopy, External Camera Photography, FA, General Microscopy, Hardcody, ICG, Intraoral Radiography, Laser Surface Scan, Magnetic Resonance Angiography, Mammography, Magnetic Resonance, Nuclear Medicine, Other, Positron Emission Tomography, Panoramic X-ray, Radio Fluoroscophy, Radiographic Imaging, Radiotherapy Dose, Radiotherapy Image, Radiotherapy Plane, Radiotherapy Record, Radiotherapy Structure Set, Red Free, Slide Microscopy, Single Photon Emission Computer Tomography, Thermography, Ultrasound, and X-ray Angiography. | | | rowspan="1" colspan="2" |Unknown Modality, Biomagnetic Imaging, Color Flow Doppler, Computed Radiography, Computed Tomography, Diaphanography, Digital Radiography, Duplex Doppler, Endoscopy, External Camera Photography, FA, General Microscopy, Hardcody, ICG, Intraoral Radiography, Laser Surface Scan, Magnetic Resonance Angiography, Mammography, Magnetic Resonance, Nuclear Medicine, Other, Positron Emission Tomography, Panoramic X-ray, Radio Fluoroscophy, Radiographic Imaging, Radiotherapy Dose, Radiotherapy Image, Radiotherapy Plane, Radiotherapy Record, Radiotherapy Structure Set, Red Free, Slide Microscopy, Single Photon Emission Computer Tomography, Thermography, Ultrasound, and X-ray Angiography. |
| − |
| |
| | |- | | |- |
| | |Image endian order | | |Image endian order |
| Line 206: |
Line 185: |
| | |- | | |- |
| | |1-5 dimension | | |1-5 dimension |
| − |
| |
| | | rowspan="1" colspan="2" |shows resolution for each dimension. | | | rowspan="1" colspan="2" |shows resolution for each dimension. |
| − |
| |
| | |- | | |- |
| | |'''Slice thickness''' | | |'''Slice thickness''' |
| | | rowspan="1" colspan="2" |shows the slice thickness. | | | rowspan="1" colspan="2" |shows the slice thickness. |
| − |
| |
| | |- | | |- |
| | |Units of measure | | |Units of measure |
| Line 218: |
Line 194: |
| | |- | | |- |
| | | rowspan="1" colspan="3" |Orientation/Origin tab | | | rowspan="1" colspan="3" |Orientation/Origin tab |
| − | | + | |- |
| − | |- | rowspan="1" colspan="3" | | + | | rowspan="1" colspan="3" |[[Image:ZeroBased_ImageAttributes3.jpg]] |
| − | [[Image:ZeroBased_ImageAttributes3.jpg]] | + | |
| − | | + | |
| | |- | | |- |
| | |Image Orientation | | |Image Orientation |
| − | | + | | rowspan="1" colspan="2" |Medical image data are stored in a variety of ways. In order to orient a two-dimensional image slice or three-dimensional volume in space, one needs to know how the order of the image data relates to a standard set of axes. For medical imaging, these axes are generally chosen to correspond to the directions '''posterior/anterior''''''superior/inferior [[http://lcni.uoregon.edu/~jolinda/MRIConvert/fileformats.htm ]]. This information could be found from the image header. To change image orientation, select one from the list: Axial, Coronal, Sagittal, or Unknown. See also "About medical image coordinate systems." |
| − | | rowspan="1" colspan="2" |Medical image data are stored in a variety of ways. In order to orient a two-dimensional image slice or three-dimensional volume in space, one needs to know how the order of the image data relates to a standard set of axes. For medical imaging, these axes are generally chosen to correspond to the directions <span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline"><nowiki>'''</nowiki><font color="#000000">posterior/anterior</font><nowiki>'''</nowiki></span>, <span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline"><nowiki>'''</nowiki><font color="#000000">superior/inferior </font><nowiki>'''</nowiki></span>and<span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline"><nowiki>'''</nowiki><font color="#000000"> left/right</font><nowiki>'''</nowiki></span><nowiki>[http://lcni.uoregon.edu/~jolinda/MRIConvert/fileformats.htm . This information could be found from the image header. To change image orientation, select one from the list: Axial, Coronal, Sagittal, or Unknown. See also "About medical image coordinate systems." ]</nowiki></div> | + | |
| − | | + | |
| | |- | | |- |
| − | | rowspan="3" colspan="1" |X-axis orientation (image left to right)</div><div class="CellBody">Y-axis orientation (image top to bottom)</div><div class="CellBody">Z-axis orientation (into the screen)</div>
| + | |X-axis orientation (image left to right)<br />Y-axis orientation (image top to bottom)<br />Z-axis orientation (into the screen)<br /> |
| − | | rowspan="3" colspan="2" |To specify X, Y, or Z -axis orientation, select one from the list: Unknown, Patient Right to Left, Patient Left to Right, Patient Anterior to Posterior, Patient Posterior to Anterior, Patient Inferior to Superior, Patient Superior to Inferior. Here, </div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 6pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: -12pt; text-transform: none; vertical-align: baseline"><font color="#000000"> The head end is referred to as the <span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline"><nowiki>'''</nowiki><font color="#000000">superior end</font><nowiki>'''</nowiki></span>, while the feet are referred to as the <span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline"><nowiki>'''</nowiki><font color="#000000">inferior end.</font><nowiki>'''</nowiki></span><br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 6pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: -12pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline"><nowiki>'''</nowiki><font color="#000000">Anterior</font><nowiki>'''</nowiki></span> refers to the "front" of the individual, similarly, <span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline"><nowiki>'''</nowiki><font color="#000000">posterior</font><nowiki>'''</nowiki></span> refers to the "back" of the individual.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 6pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: -12pt; text-transform: none; vertical-align: baseline"><font color="#000000"> Left and right are self explanatory. <br /></font></div> | + | | rowspan="1" colspan="2" |To specify X, Y, or Z -axis orientation, select one from the list: Unknown, Patient Right to Left, Patient Left to Right, Patient Anterior to Posterior, Patient Posterior to Anterior, Patient Inferior to Superior, Patient Superior to Inferior. Here, the head end is referred to as the '''superior end''', while the feet are referred to as the '''inferior end.'''<br />'''Anterior'''refers to the "front" of the individual, similarly, '''posterior''' refers to the "back" of the individual.<br />Left and right are self explanatory. |
| − | | + | |
| | |- | | |- |
| − | | | + | |X-axis origin<br /> Y-axis origin<br />Z-axis origin<br /> |
| − | | + | |
| − | <div class="CellBody">X-axis origin</div><div class="CellBody">Y-axis origin</div><div class="CellBody">Z-axis origin</div>
| + | |
| − | | + | |
| | | rowspan="1" colspan="2" |The image origin is generally at the corner of the image, often the upper left, the axis directions are: the X- axis is across, Y -axis is down, and Z-axis goes into the screen. Usually, it differs from matrix coordinates and Cartesian coordinates. E.g. for the above image, the origin coordinates shown in the dialog box are as follows: x=0,z=0, and y=-239.0625, which corresponds to the scanner frame of reference and scanner origin coordinates. If you mouse over the image, MIPAV will show you the coordinates from the image frame of reference. | | | rowspan="1" colspan="2" |The image origin is generally at the corner of the image, often the upper left, the axis directions are: the X- axis is across, Y -axis is down, and Z-axis goes into the screen. Usually, it differs from matrix coordinates and Cartesian coordinates. E.g. for the above image, the origin coordinates shown in the dialog box are as follows: x=0,z=0, and y=-239.0625, which corresponds to the scanner frame of reference and scanner origin coordinates. If you mouse over the image, MIPAV will show you the coordinates from the image frame of reference. |
| − |
| |
| | |- | | |- |
| | |4-th dimension | | |4-th dimension |
| | | rowspan="1" colspan="2" |For 4D images, it specifies the units and values used for the 4-th dimension. | | | rowspan="1" colspan="2" |For 4D images, it specifies the units and values used for the 4-th dimension. |
| − |
| |
| | |- | | |- |
| | | rowspan="1" colspan="3" |Transform matrix | | | rowspan="1" colspan="3" |Transform matrix |
| − |
| |
| | |- | | |- |
| | | rowspan="1" colspan="3" | | | | rowspan="1" colspan="3" | |
| | [[Image:ImageAttributesTransformMatrix1.jpg]] | | [[Image:ImageAttributesTransformMatrix1.jpg]] |
| − |
| |
| | |- | | |- |
| | |Matrix | | |Matrix |
| | | rowspan="1" colspan="2" |displays a list of transformation matrix for the chosen image. | | | rowspan="1" colspan="2" |displays a list of transformation matrix for the chosen image. |
| − |
| |
| | |- | | |- |
| | |Transform. ID | | |Transform. ID |
| | | rowspan="1" colspan="2" |displays transformation ID type for the chosen image. To change the current transformation matrix, select a new transform ID first, and then click Replace. This will replace the transformation matrix. | | | rowspan="1" colspan="2" |displays transformation ID type for the chosen image. To change the current transformation matrix, select a new transform ID first, and then click Replace. This will replace the transformation matrix. |
| − |
| |
| | |- | | |- |
| | |Replace | | |Replace |
| | | rowspan="1" colspan="2" |replaces a current transformation matrix to a new one that you choose. | | | rowspan="1" colspan="2" |replaces a current transformation matrix to a new one that you choose. |
| − |
| |
| | |- | | |- |
| | |Add a New | | |Add a New |
| | | rowspan="1" colspan="2" |adds a transformation matrix that has been selected from the Transform ID list to the image. | | | rowspan="1" colspan="2" |adds a transformation matrix that has been selected from the Transform ID list to the image. |
| − |
| |
| | |- | | |- |
| | |Rename | | |Rename |
| | | rowspan="1" colspan="2" |renames a transformation matrix. | | | rowspan="1" colspan="2" |renames a transformation matrix. |
| − |
| |
| | |- | | |- |
| | |Copy | | |Copy |
| | | rowspan="1" colspan="2" |copies the current transformation matrix to the Clipboard. | | | rowspan="1" colspan="2" |copies the current transformation matrix to the Clipboard. |
| − |
| |
| | |- | | |- |
| | |Paste | | |Paste |
| | | rowspan="1" colspan="2" |pastes a copied transformation matrix | | | rowspan="1" colspan="2" |pastes a copied transformation matrix |
| − |
| |
| | |- | | |- |
| | |Load | | |Load |
| | | rowspan="1" colspan="2" |loads a transformation matrix (from a file with the ''*.mtx'' | | | rowspan="1" colspan="2" |loads a transformation matrix (from a file with the ''*.mtx'' |
| − |
| |
| | |- | | |- |
| | |Save | | |Save |
| | | rowspan="1" colspan="2" |saves a transformation matrix to a file. | | | rowspan="1" colspan="2" |saves a transformation matrix to a file. |
| − |
| |
| | |- | | |- |
| | |Identity | | |Identity |
| − |
| |
| | | rowspan="1" colspan="2" |transforms the current transformation matrix to an identity matrix. | | | rowspan="1" colspan="2" |transforms the current transformation matrix to an identity matrix. |
| − |
| |
| | |- | | |- |
| | |Invert | | |Invert |
| | | rowspan="1" colspan="2" |inverts the current transformation matrix. | | | rowspan="1" colspan="2" |inverts the current transformation matrix. |
| − |
| |
| | |- | | |- |
| | |Composite | | |Composite |
| | | rowspan="1" colspan="2" |TBD. | | | rowspan="1" colspan="2" |TBD. |
| − |
| |
| | |- | | |- |
| | |Decompose | | |Decompose |
| − |
| |
| | | rowspan="1" colspan="2" |TBD. | | | rowspan="1" colspan="2" |TBD. |
| − | | + | |- |
| − | |- | rowspan="1" colspan="3" |Talairach | + | | rowspan="1" colspan="3" |Talairach |
| − | | + | |- |
| − | |- | | + | |Shows ACPC and Talairach transformation matrices. |
| − | | + | |
| − | <div class="CellBody">Shows ACPC and Talairach transformation matrices</div>
| + | |
| − | | + | |
| | | rowspan="12" colspan="2" | | | | rowspan="12" colspan="2" | |
| | [[Image:ImageAttributesTalairach1.jpg]] | | [[Image:ImageAttributesTalairach1.jpg]] |
| | |- | | |- |
| | |Include Talairach - displays a Talairach transformation matrix for the image. | | |Include Talairach - displays a Talairach transformation matrix for the image. |
| − |
| |
| | |- | | |- |
| | |Load -loads a new matrix. | | |Load -loads a new matrix. |
| − |
| |
| | |- | | |- |
| | |Save - saves a matrix to a file. | | |Save - saves a matrix to a file. |
| − |
| |
| | |- | | |- |
| | | rowspan="8" colspan="1" | | | | rowspan="8" colspan="1" | |
| | + | |- |- |- |- |- |- |- |-} |
| | + | |} |
| | + | |
| | | | |
| − | <div class="CellBody"> </div>
| |
| | | | |
| − | |- |- |- |- |- |- |- |-}
| |
| | | | |
| − | ==testing html2wiki==
| |
| − | !DOCTYPE HTML PUBLIC "-//W3C//DTD HTML 4.0 Transitional//EN" "http://www.w3.org/TR/REC-html40/loose.dtd"><html><head><meta http-equiv="Content-Type" content="text/html; charset=ISO-8859-1"><meta name="GENERATOR" content="Quadralay WebWorks Publisher Standard Edition 7.0.6.1257"><meta name="TEMPLATEBASE" content="Dynamic HTML Standard Edition"><meta name="LASTUPDATED" content="02/07/12 12:25:06"><link rel="StyleSheet" href="standard.css" type="text/css" media="screen"><title>Supported graphic formats overview </title></head><body background="images/backgrnd.gif">
| |
| − | <img src="images/wwplogo.gif" width="122" height="63" alt="Put your logo here!"><hr align="left"><blockquote><div style="color: #000000; font-style: normal; font-weight: bold; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 24pt; text-align: right; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1159719"></a>4
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: bold; margin-bottom: 600pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: right; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1159723"></a>Understanding Image Basics
| |
| − | </div><div style="color: #0023a9; font-style: normal; font-weight: bold; margin-bottom: 6pt; margin-left: 6pt; margin-right: 6pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220216"></a><span style="color: #0023a9; font-style: normal; font-weight: bold; text-decoration: none; text-transform: none; vertical-align: baseline">In this </span>chapter<span style="color: #0023a9; font-style: normal; font-weight: bold; text-decoration: none; text-transform: none; vertical-align: baseline"> . . .</span>
| |
| − | </div><div style="color: #0023a9; font-style: normal; font-weight: normal; margin-bottom: 1pt; margin-left: 18pt; margin-right: 6pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: -12pt; text-transform: none; vertical-align: baseline"><a name="1220220"></a><a href="MIPAV_ImageBasics.html#1220264">"Supported graphic formats overview" on page 136</a>
| |
| − | </div><div style="color: #0023a9; font-style: normal; font-weight: normal; margin-bottom: 1pt; margin-left: 18pt; margin-right: 6pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: -12pt; text-transform: none; vertical-align: baseline"><a name="1220223"></a><a href="MIPAV_ImageBasics.html#1220316">"Understanding the MIPAV dimensionality and coordinate system" on page 136</a>
| |
| − | </div><div style="color: #0023a9; font-style: normal; font-weight: normal; margin-bottom: 1pt; margin-left: 18pt; margin-right: 6pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: -12pt; text-transform: none; vertical-align: baseline"><a name="1220228"></a><a href="MIPAV_ImageBasics.html#1220350">"Data types" on page 138</a>
| |
| − | </div><div style="color: #0023a9; font-style: normal; font-weight: normal; margin-bottom: 1pt; margin-left: 18pt; margin-right: 6pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: -12pt; text-transform: none; vertical-align: baseline"><a name="1220232"></a><a href="MIPAV_ImageBasics.html#1220413">"Headers" on page 138</a>
| |
| − | </div><div style="color: #0023a9; font-style: normal; font-weight: normal; margin-bottom: 1pt; margin-left: 18pt; margin-right: 6pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: -12pt; text-transform: none; vertical-align: baseline"><a name="1220236"></a><a href="MIPAV_ImageBasics.html#1220437">"Endianness" on page 139</a>
| |
| − | </div><div style="color: #0023a9; font-style: normal; font-weight: normal; margin-bottom: 1pt; margin-left: 6pt; margin-right: 6pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220240"></a><a href="MIPAV_ImageBasics.html#1220445">"Viewing and editing image attributes" on page 139</a>
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220241"></a>
| |
| − | </div><p class="Body"><a name="1220245"></a>This chapter also explains the dimensionality and coordinate system used by MIPAV, the data types MIPAV supports, the header offset information, endianness, and how to view and edit image attributes.It also provides a basic information about image file formats supported by MIPAV. More information about graphic file formats and the formats that MIPAV supports can be found in Appendix C: "Supported formats" .
| |
| − | <h2 class="Heading1"><a name="1220264"></a>Supported graphic formats overview
| |
| − | </h2><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220268"></a>MIPAV supports three categories of graphic file formats: vector (i.e., volume of interest, or VOI), bitmap (i.e., 2D to 5D grayscale or RGB images), and 3D images.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220270"></a><span style="color: #000000; font-style: normal; font-weight: bold; text-decoration: none; text-transform: none; vertical-align: baseline">Vector file formats </span>are usually used to store line-based elements. These elements can be geometric shapes, such as polygons, curves, or splines. MIPAV can read and generate several vector-formatted file types including the MIPAV lookup table (LUT), MIPAV MTX, MIPAV Graphics Plot (PLT), and MIPAV VOI. See also Appendix C: "Supported formats" on page 561.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220290"></a><span style="color: #000000; font-style: normal; font-weight: bold; text-decoration: none; text-transform: none; vertical-align: baseline">Bitmap file formats</span> are usually used to store real-world graphics data e.g., photographs or medical images such as CT scans. The data portion of bitmap files contains numerical data that indicates the color of each pixel or voxel in the image. MIPAV can read and generate a number of bitmap formatted file types, such as Adobe Photoshop (PSD), Graphics Interchange File (GIF), Sun Raster (RS), Tagged Image File Format (TIFF), and Truevision Graphics Adapter (TGA).
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220297"></a><span style="color: #000000; font-style: normal; font-weight: bold; text-decoration: none; text-transform: none; vertical-align: baseline">MIPAV uses 3D graphic formatted files </span>to store descriptions of the color and shape of 3D models of real-world and imaginary objects.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220300"></a>To support the wide range of image-processing needs of the NIH intramural research community, MIPAV reads and writes the image file types indicated in Table 1, refer to Appendix C: "Supported formats" on page 561.
| |
| − | </div><h2 class="Heading1"><a name="1220316"></a>Understanding the MIPAV dimensionality and coordinate system
| |
| − | </h2><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220321"></a>Internally, MIPAV is designed to store images of any dimensionality and thus is <span style="color: #000000; font-style: italic; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">n </span>dimensional. <a href="MIPAV_ImageBasics.html#1220345">Figure 59</a> is a model of an example 3D-image where each position within the grid represents a <span style="color: #000000; font-style: italic; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">voxel</span>.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 5pt; margin-left: 0pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220328"></a><span style="color: #000000; font-style: normal; font-weight: bold; text-decoration: none; text-transform: none; vertical-align: baseline">voxel</span>-The smallest distinguishable cube-shaped part of a 3D-image.
| |
| − | </div><p class="Body"><a name="1220329"></a>The voxel in the upper left-most position is considered the origin of the dataset. (A volumetric dataset is a set of cross-sectional images). Ideally, each voxel would be isotropic (i.e., cubic), but clinical datasets are often acquired in such a manner that the resolution in the <span style="color: #000000; font-style: italic; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">x</span> and <span style="color: #000000; font-style: italic; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">y</span> dimensions is greater than the resolution in the <span style="color: #000000; font-style: italic; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">z</span> dimension, thus producing rectangular voxels (elongated in the <span style="color: #000000; font-style: italic; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">z</span> direction). This voxel anisotropy is taken into account in the execution of the algorithms found in MIPAV.
| |
| | | | |
| − | <p class="Body"><a name="1220336"></a>Because MIPAV's image storage buffer is <span style="color: #000000; font-style: italic; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">n</span>-dimensional, it can store 2D, 3D, and 4D+ medical images. CT and MRI are examples of 3D medical images where the three dimensions (<span style="color: #000000; font-style: italic; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">x, y,</span> and <span style="color: #000000; font-style: italic; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">z</span>) are spatial dimensions. Other 3D datasets include fluoroscopy volumes, where the first two dimensions are spatial and the third dimension is time.
| |
| − | <div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220346"></a>
| |
| − | {| border="1" cellpadding="5" cellspacing="0"
| |
| − | |+<div style="color: #000000; font-style: normal; font-weight: bold; margin-bottom: 3pt; margin-left: 0pt; margin-right: 0pt; margin-top: 9pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220345"></a>Figure 59. Example of 3D image (origin at top left)
| |
| − | </div>
| |
| − | |-
| |
| − | | <div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220342"></a><br clear="all" />
| |
| − | {| align="center"
| |
| − | | <img src="images/MIPAV_ImageBasicsa.gif" height="470" width="568" align="center" border="0" hspace="0" vspace="0">
| |
| − | |}<br clear="all">
| |
| − | </div>
| |
| − | |}
| |
| − | </div><h2 class="Heading1"><a name="1220350"></a>Data types
| |
| − | </h2><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220351"></a>A <span style="color: #000000; font-style: italic; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">data type</span> is the classification of a particular type of information. For example, a floating point data type indicates a number with a decimal point. Data read from image files are stored in MIPAV's data structures. MIPAV supports all the basic data types shown in <a href="MIPAV_ImageBasics.html#1220361">Table 1</a> and may in the future support more composite data types including color and complex numbers. The image data read from the file is stored in a data buffer of the same basic data type, preventing the reduction of image information that results from conversion into a fixed data type.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220408"></a>
| |
| − | {| border="1" cellpadding="5" cellspacing="0"
| |
| − | |+<a name="1220361"></a><div class="TableTitle">Table 1. Data types supported by MIPAV</div>
| |
| − | |-
| |
| − | | <a name="1220365"></a><div class="CellHeading">Data type</div>
| |
| − | | <a name="1220367"></a><div class="CellHeading">Description</div>
| |
| − | |-
| |
| − | | <a name="1220369"></a><div class="CellBody">Boolean</div>
| |
| − | | <a name="1220371"></a><div class="CellBody">1 bit per pixel/voxel (1 on, 0 off)</div>
| |
| − | |-
| |
| − | | <a name="1220373"></a><div class="CellBody">Signed byte</div>
| |
| − | | <a name="1220375"></a><div class="CellBody">1 byte per pixel/voxel (-128, 127)</div>
| |
| − | |-
| |
| − | | <a name="1220377"></a><div class="CellBody">Unsigned byte</div>
| |
| − | | <a name="1220379"></a><div class="CellBody">1 byte per pixel/voxel (0, 255)</div>
| |
| − | |-
| |
| − | | <a name="1220381"></a><div class="CellBody">Signed short</div>
| |
| − | | <a name="1220383"></a><div class="CellBody">2 bytes per pixel/voxel (-32768, 32767)</div>
| |
| − | |-
| |
| − | | <a name="1220385"></a><div class="CellBody">Unsigned short</div>
| |
| − | | <a name="1220387"></a><div class="CellBody">2 bytes per pixel/voxel (0, 65535)</div>
| |
| − | |-
| |
| − | | <a name="1220389"></a><div class="CellBody">Integer</div>
| |
| − | | <a name="1220391"></a><div class="CellBody">4 bytes per pixel/voxel (-2<span style="color: #000000; font-style: normal; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: super">31</span>, 2<span style="color: #000000; font-style: normal; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: super">31</span>-1)</div>
| |
| − | |-
| |
| − | | <a name="1220393"></a><div class="CellBody">Long</div>
| |
| − | | <a name="1220395"></a><div class="CellBody">8 bytes per pixel/voxel (-9.22E18, 9.22E18)</div>
| |
| − | |-
| |
| − | | <a name="1220397"></a><div class="CellBody">Float</div>
| |
| − | | <a name="1220399"></a><div class="CellBody">4 bytes per pixel/voxel (-3.4E38, 3.4E38)</div>
| |
| − | |-
| |
| − | | <a name="1220401"></a><div class="CellBody">Double</div>
| |
| − | | <a name="1220403"></a><div class="CellBody">8 bytes per pixel/voxel (-1.8E308, 1.8E308)</div>
| |
| − | |-
| |
| − | | <a name="1220405"></a><div class="CellBody">Color 32</div>
| |
| − | | <a name="1220407"></a><div class="CellBody">3 bytes per pixel/voxel, plus 1 byte; 8 bits per color channel (alpha, red, green, and blue)</div>
| |
| − | |}
| |
| − | </div><h2 class="Heading1"><a name="1220413"></a>Headers
| |
| − | </h2><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220417"></a>If you attempt to open a raw image, you may need to supply MIPAV with the <span style="color: #000000; font-style: italic; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">header offset.</span> The header offset indicates the size of the space reserved at the beginning of the file where specific types of information is kept. This space, which is called the header, precedes the image data. If you know the length of the header, enter it in the header offset text box. When MIPAV accesses the file, it skips the header offset and begins to read the image data. <span style="color: #000000; font-style: normal; font-weight: bold; text-decoration: none; text-transform: none; vertical-align: baseline">Note that</span> not all image file formats have a header.
| |
| − | </div><h2 class="Heading1"><a name="1220437"></a>Endianness
| |
| − | </h2><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220438"></a><span style="color: #000000; font-style: italic; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">Endianness</span> refers to the byte ordering of the data. Some computers order the data with the least significant byte (LSB) first followed by the most significant byte (MSB). This byte order is referred as "little endian" or Intel byte ordering. machines that use little-endian byte ordering are VAXes, Intel x86, and Pentium. The reverse is MSB then LSB, which is referred as "big endian" or Motorola byte ordering. Machines that use big-endian byte ordering are IBM System 3D, RISC, and a Motorola 680x0. MIPAV is biendian; that is, it supports both big- and little-endian byte-ordering formats.
| |
| − | </div><h2 class="Heading1"><a name="1220445"></a>Viewing and editing image attributes
| |
| | | | |
| − | </h2><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220446"></a>Information about the image appears in the Image Information dialog box and in the Image Attributes dialog box. | + | |
| − | </div><p class="Body"><a name="1220447"></a>The Image Attributes window displays the name of the image file and information about the resolution of the image. The About Image window displays more detailed information about the image, such as the orientation of the image or the number of bits per pixel or voxel.
| + | |
| − | <h3 class="Heading2"><a name="1220452"></a>Viewing image attributes
| + | |
| − | </h3><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1221044"></a>To view the attributes of an image, select Image > Attributes > View Header in the MIPAV window. For DICOM datasets, the Image Information dialog box similar to the one shown in <a href="MIPAV_ImageBasics.html#1221393">Figure 60</a>-a appears. For non-DICOM datasets, an Image Information dialog box similar to the one in <a href="MIPAV_ImageBasics.html#1221393">Figure 60</a>-b appears.
| + | |
| − | </div><p class="Body"><a name="1221323"></a><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1221329"></a>
| + | |
| − | {| border="1" cellpadding="5" cellspacing="0"
| + | |
| − | |+<div style="color: #000000; font-style: normal; font-weight: bold; margin-bottom: 3pt; margin-left: 0pt; margin-right: 0pt; margin-top: 9pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1221393"></a>Figure 60. The Info dialog box for DICOM (a) and non-DICOM (b) images
| + | <br /> |
| − | </div>
| + | '''To modify the name of the image dataset<br />''' |
| − | |-
| + | 1 Open either a DICOM or non-DICOM image dataset.<br /> |
| − | | <div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 1pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1221385"></a><br clear="all" />
| + | 2 Select Image > Attributes > Edit Attributes in the MIPAV window. The Image Attributes dialog box opens.<br /> |
| − | {| align="center"
| + | 3 Select the name of the image in the Image name (without suffix) box.<br /> |
| − | | <img src="images/ImageBasicsInfoDICOMDialog.jpg" height="457" width="501" align="center" border="0" hspace="0" vspace="0">
| + | 4 Type a new name of the image. <br /> |
| − | |}<br clear="all">
| + | 5 Click either Apply or OK. The new image name appears in the header on the image window.<br /> |
| − | </div>
| + | |
| − | |-
| + | |
| − | | <a name="1221387"></a><div class="CellBody">(a) the Info dialog box for DICOM images</div>
| + | {| border="1" cellpadding="5" |
| − | |-
| + | |
| − | | <div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 1pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1221389"></a><br clear="all" />
| + | |
| − | {| align="center"
| + | |
| − | | <img src="images/ImageBasicsInfoDialog.jpg" height="340" width="501" align="center" border="0" hspace="0" vspace="0">
| + | |
| − | |}<br clear="all">
| + | |
| − | </div>
| + | |
| − | |-
| + | |
| − | | <a name="1221391"></a><div class="CellBody">(b) the Info dialog box for non-DICOM images</div>
| + | |
| − | |}
| + | |
| − | </div><h3 class="Heading2"><a name="1221249"></a>Editing image attributes
| + | |
| − | </h3><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220478"></a>You can edit some of the image attributes for both DICOM datasets and non-DICOM datasets in the Image Attributes dialog box (<a href="MIPAV_ImageBasics.html#1223078">Figure 61</a>). The Image Attributes dialog box includes five tabbed pages, which are the following:
| + | |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><a name="1220482"></a><span style="color: #000000; font-style: normal; font-weight: bold; text-decoration: none; text-transform: none; vertical-align: baseline">General page</span> displays the name of the image (without its extension, or suffix) and lists the image modality
| + | |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><a name="1220483"></a><span style="color: #000000; font-style: normal; font-weight: bold; text-decoration: none; text-transform: none; vertical-align: baseline">Resolutions page</span> indicates the unit of measure for each dimension | + | |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><a name="1220484"></a><span style="color: #000000; font-style: normal; font-weight: bold; text-decoration: none; text-transform: none; vertical-align: baseline">Orientations/Origin page</span> lists the image orientations
| + | |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><a name="1220486"></a><span style="color: #000000; font-style: normal; font-weight: bold; text-decoration: none; text-transform: none; vertical-align: baseline">Transform Matrix</span> displays and lists the values for the transform matrix | + | |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><a name="1221510"></a><span style="color: #000000; font-style: normal; font-weight: bold; text-decoration: none; text-transform: none; vertical-align: baseline">Talairach</span> displays Talairach transform
| + | |
| − | </div><p class="Body"><a name="1222859"></a>By default, the General page always appears first when the Image Attributes dialog box opens.
| + | |
| − | <h4 class="Heading3"><a name="1223159"></a>Image Attributes dialog box
| + | |
| − | </h4><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1223083"></a>
| + | |
| − | {| border="1" cellpadding="5" cellspacing="0" | + | |
| | |+ | | |+ |
| − | |- | + | |- |
| − | | <a name="1223698"></a><div class="CellBody">Apply</div> | + | | |
| − | | colspan="2" rowspan="1" | <a name="1223700"></a><div class="CellBody">applies the changes to the current image, but doesn't close the dialog box.</div>
| + | <div style="font-style: normal; margin-bottom: 1pt; margin-left: 0pt; margin-right: 0pt; margin-top: 17pt; text-align: center; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline">'''<font color="#000000"> What's the Difference Between<br />Clicking Apply or Clicking OK?<br /></font>'''</div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 7pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> Click Apply when you want the dialog box or window to remain open after it makes the change so that you can make further modifications. If instead you click OK, the dialog box or window closes immediately after making the current change.<br /></font></div> |
| − | |-
| + | |
| − | | <a name="1223704"></a><div class="CellBody">OK</div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1223706"></a><div class="CellBody">applies the changes to the current image and closes the dialog box.</div>
| + | |
| − | |-
| + | |
| − | | <a name="1223710"></a><div class="CellBody">Close</div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1223712"></a><div class="CellBody">closes the dialog box without applying the changes to the image.</div>
| + | |
| − | |-
| + | |
| − | | <a name="1223716"></a><div class="CellBody">Help</div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1223718"></a><div class="CellBody">opens the on-line help.</div>
| + | |
| − | |-
| + | |
| − | | colspan="3" rowspan="1" | <a name="1222862"></a><div class="CellBody"><span style="color: #000000; font-style: normal; font-weight: bold; text-decoration: none; text-transform: none; vertical-align: baseline">General tab</span></div>
| + | |
| − | |-
| + | |
| − | | colspan="3" rowspan="1" | <div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1222871"></a><br clear="all" />
| + | |
| − | {| align="center"
| + | |
| − | | <img src="images/ImageAttributesGeneral.jpg" height="338" width="647" align="center" border="0" hspace="0" vspace="0">
| + | |
| − | |}<br clear="all">
| + | |
| − | </div>
| + | |
| − | |-
| + | |
| − | | <a name="1222877"></a><div class="CellBody">Image name (without suffix)</div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1222879"></a><div class="CellBody">displays the name of the current image. You can change the image's name here.</div>
| + | |
| − | |-
| + | |
| − | | <a name="1222883"></a><div class="CellBody">Image modality</div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1222885"></a><div class="CellBody">shows image modality. You can change the modality by selecting it from the list.</div>
| + | |
| − | |-
| + | |
| − | | <a name="1222889"></a><div class="CellBody"> The list includes: </div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1222891"></a><div class="CellBody">Unknown Modality, Biomagnetic Imaging, Color Flow Doppler, Computed Radiography, Computed Tomography, Diaphanography, Digital Radiography, Duplex Doppler, Endoscopy, External Camera Photography, FA, General Microscopy, Hardcody, ICG, Intraoral Radiography, Laser Surface Scan, Magnetic Resonance Angiography, Mammography, Magnetic Resonance, Nuclear Medicine, Other, Positron Emission Tomography, Panoramic X-ray, Radio Fluoroscophy, Radiographic Imaging, Radiotherapy Dose, Radiotherapy Image, Radiotherapy Plane, Radiotherapy Record, Radiotherapy Structure Set, Red Free, Slide Microscopy, Single Photon Emission Computer Tomography, Thermography, Ultrasound, and X-ray Angiography. </div>
| + | |
| − | |-
| + | |
| − | | <a name="1222895"></a><div class="CellBody">Image endian order</div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1222897"></a><div class="CellBody">Here, use the appropriate radio button to set the image endiannes. </div>
| + | |
| − | |-
| + | |
| − | | colspan="3" rowspan="1" | <a name="1222901"></a><div class="CellBody">Resolutions tab</div>
| + | |
| − | |-
| + | |
| − | | colspan="3" rowspan="1" | <div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1222910"></a><br clear="all" />
| + | |
| − | {| align="center"
| + | |
| − | | <img src="images/ImageAttributesDialogREsolutionsTab.jpg" height="380" width="700" align="center" border="0" hspace="0" vspace="0">
| + | |
| − | |}<br clear="all">
| + | |
| − | </div>
| + | |
| − | |-
| + | |
| − | | <a name="1222916"></a><div class="CellBody">1-5 dimension</div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1222918"></a><div class="CellBody">shows resolution for each dimension.</div>
| + | |
| − | |-
| + | |
| − | | <a name="1222922"></a><div class="CellBody"><span style="color: #000000; font-style: normal; font-weight: bold; text-decoration: none; text-transform: none; vertical-align: baseline">Slice thickness</span></div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1222924"></a><div class="CellBody">shows the slice thickness.</div>
| + | |
| − | |-
| + | |
| − | | <a name="1222928"></a><div class="CellBody">Units of measure</div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1222930"></a><div class="CellBody">displays the units of measure used in the image.</div>
| + | |
| − | |-
| + | |
| − | | colspan="3" rowspan="1" | <a name="1222934"></a><div class="CellBody">Orientation/Origin tab</div>
| + | |
| − | |-
| + | |
| − | | colspan="3" rowspan="1" | <div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1222943"></a><br clear="all" />
| + | |
| − | {| align="center"
| + | |
| − | | <img src="images/ZeroBased_ImageAttributes3.jpg" height="389" width="647" align="center" border="0" hspace="0" vspace="0">
| + | |
| − | |}<br clear="all">
| + | |
| − | </div>
| + | |
| − | |-
| + | |
| − | | <a name="1222949"></a><div class="CellBody">Image Orientation</div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1222951"></a><div class="CellBody">Medical image data are stored in a variety of ways. In order to orient a two-dimensional image slice or three-dimensional volume in space, one needs to know how the order of the image data relates to a standard set of axes. For medical imaging, these axes are generally chosen to correspond to the directions <span style="color: #000000; font-style: normal; font-weight: bold; text-decoration: none; text-transform: none; vertical-align: baseline">posterior/anterior</span>, <span style="color: #000000; font-style: normal; font-weight: bold; text-decoration: none; text-transform: none; vertical-align: baseline">superior/inferior </span>and<span style="color: #000000; font-style: normal; font-weight: bold; text-decoration: none; text-transform: none; vertical-align: baseline"> left/right</span><a href="http://lcni.uoregon.edu/~jolinda/MRIConvert/fileformats.htm">. This information could be found from the image header. To change image orientation, select one from the list: Axial, Coronal, Sagittal, or Unknown. See also "About medical image coordinate systems." </a></div>
| + | |
| − | |-
| + | |
| − | | colspan="1" rowspan="3" | <a name="1222956"></a><div class="CellBody">X-axis orientation (image left to right)</div><a name="1222957"></a><div class="CellBody">Y-axis orientation (image top to bottom)</div><a name="1222958"></a><div class="CellBody">Z-axis orientation (into the screen)</div>
| + | |
| − | | colspan="2" rowspan="3" | <a name="1222960"></a><div class="CellBody">To specify X, Y, or Z -axis orientation, select one from the list: Unknown, Patient Right to Left, Patient Left to Right, Patient Anterior to Posterior, Patient Posterior to Anterior, Patient Inferior to Superior, Patient Superior to Inferior. Here, </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 6pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: -12pt; text-transform: none; vertical-align: baseline"><a name="1222961"></a> The head end is referred to as the <span style="color: #000000; font-style: normal; font-weight: bold; text-decoration: none; text-transform: none; vertical-align: baseline">superior end</span>, while the feet are referred to as the <span style="color: #000000; font-style: normal; font-weight: bold; text-decoration: none; text-transform: none; vertical-align: baseline">inferior end.</span>
| + | |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 6pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: -12pt; text-transform: none; vertical-align: baseline"><a name="1222962"></a><span style="color: #000000; font-style: normal; font-weight: bold; text-decoration: none; text-transform: none; vertical-align: baseline">Anterior</span> refers to the "front" of the individual, similarly, <span style="color: #000000; font-style: normal; font-weight: bold; text-decoration: none; text-transform: none; vertical-align: baseline">posterior</span> refers to the "back" of the individual.
| + | |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 6pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: -12pt; text-transform: none; vertical-align: baseline"><a name="1222963"></a> Left and right are self explanatory.
| + | |
| − | </div>
| + | |
| − | |-
| + | |
| − | |-
| + | |
| − | |-
| + | |
| − | | <a name="1222979"></a><div class="CellBody">X-axis origin</div><a name="1222980"></a><div class="CellBody">Y-axis origin</div><a name="1222981"></a><div class="CellBody">Z-axis origin</div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1222983"></a><div class="CellBody">The image origin is generally at the corner of the image, often the upper left, the axis directions are: the X- axis is across, Y -axis is down, and Z-axis goes into the screen. Usually, it differs from matrix coordinates and Cartesian coordinates. E.g. for the above image, the origin coordinates shown in the dialog box are as follows: x=0,z=0, and y=-239.0625, which corresponds to the scanner frame of reference and scanner origin coordinates. If you mouse over the image, MIPAV will show you the coordinates from the image frame of reference.</div>
| + | |
| − | |-
| + | |
| − | | <a name="1222987"></a><div class="CellBody">4-th dimension</div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1222989"></a><div class="CellBody">For 4D images, it specifies the units and values used for the 4-th dimension.</div>
| + | |
| − | |-
| + | |
| − | | colspan="3" rowspan="1" | <a name="1222993"></a><div class="CellBody">Transform matrix</div>
| + | |
| − | |-
| + | |
| − | | colspan="3" rowspan="1" | <div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1223002"></a><br clear="all" />
| + | |
| − | {| align="center"
| + | |
| − | | <img src="images/ImageAttributesTransformMatrix1.jpg" height="338" width="647" align="center" border="0" hspace="0" vspace="0">
| + | |
| − | |}<br clear="all">
| + | |
| − | </div>
| + | |
| − | |-
| + | |
| − | | <a name="1223008"></a><div class="CellBody">Matrix</div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1223010"></a><div class="CellBody">displays a list of transformation matrix for the chosen image </div>
| + | |
| − | |-
| + | |
| − | | <a name="1223496"></a><div class="CellBody">Transform. ID</div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1223498"></a><div class="CellBody">displays transformation ID type for the chosen image. To change the current transformation matrix, select a new transform ID first, and then click Replace. This will replace the transformation matrix.</div>
| + | |
| − | |-
| + | |
| − | | <a name="1223502"></a><div class="CellBody">Replace</div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1223504"></a><div class="CellBody">replaces a current transformation matrix to a new one that you choose.</div>
| + | |
| − | |-
| + | |
| − | | <a name="1223586"></a><div class="CellBody">Add a New</div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1223588"></a><div class="CellBody">adds a transformation matrix that has been selected from the Transform ID list to the image.</div>
| + | |
| − | |-
| + | |
| − | | <a name="1223508"></a><div class="CellBody">Rename</div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1223510"></a><div class="CellBody">renames a transformation matrix.</div>
| + | |
| − | |-
| + | |
| − | | <a name="1223514"></a><div class="CellBody">Copy</div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1223516"></a><div class="CellBody">copies the current transformation matrix to the Clipboard.</div>
| + | |
| − | |-
| + | |
| − | | <a name="1223520"></a><div class="CellBody">Paste</div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1223522"></a><div class="CellBody">pastes a copied transformation matrix.</div>
| + | |
| − | |-
| + | |
| − | | <a name="1223526"></a><div class="CellBody">Load</div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1223528"></a><div class="CellBody">loads a transformation matrix (from a file with the <span style="color: #000000; font-style: italic; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">*.mtx</span> extension)</div>
| + | |
| − | |-
| + | |
| − | | <a name="1223532"></a><div class="CellBody">Save</div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1223534"></a><div class="CellBody">saves a transformation matrix to a file.</div>
| + | |
| − | |-
| + | |
| − | | <a name="1223538"></a><div class="CellBody">Identity</div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1223540"></a><div class="CellBody">transforms the current transformation matrix to an identity matrix.</div>
| + | |
| − | |-
| + | |
| − | | <a name="1223544"></a><div class="CellBody">Invert</div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1223546"></a><div class="CellBody">inverts the current transformation matrix.</div>
| + | |
| − | |-
| + | |
| − | | <a name="1223550"></a><div class="CellBody">Composite</div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1223552"></a><div class="CellBody">TBD.</div>
| + | |
| − | |-
| + | |
| − | | <a name="1223556"></a><div class="CellBody">Decompose</div>
| + | |
| − | | colspan="2" rowspan="1" | <a name="1223558"></a><div class="CellBody">TBD.</div>
| + | |
| − | |-
| + | |
| − | | colspan="3" rowspan="1" | <a name="1223020"></a><div class="CellBody">Talairach</div>
| + | |
| − | |-
| + | |
| − | | <a name="1223628"></a><div class="CellBody">Shows ACPC and Talairach transformation matrices</div>
| + | |
| − | | colspan="2" rowspan="12" | <div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1223630"></a><br clear="all" />
| + | |
| − | {| align="center"
| + | |
| − | | <img src="images/ImageAttributesTalairach1.jpg" height="709" width="647" align="center" border="0" hspace="0" vspace="0">
| + | |
| − | |}<br clear="all">
| + | |
| − | </div>
| + | |
| − | |-
| + | |
| − | | <a name="1223622"></a><div class="CellBody">Include Talairach - <span style="color: #000000; font-style: normal; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">displays a Talairach transformation matrix for the image.</span></div>
| + | |
| − | |-
| + | |
| − | | <a name="1223616"></a><div class="CellBody">Load -<span style="color: #000000; font-style: normal; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline"> loads a new matrix.</span></div>
| + | |
| − | |-
| + | |
| − | | <a name="1223610"></a><div class="CellBody">Save - <span style="color: #000000; font-style: normal; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">saves a matrix to a file.</span></div>
| + | |
| − | |-
| + | |
| − | | colspan="1" rowspan="8" | <a name="1223065"></a><div class="CellBody"> </div>
| + | |
| − | |-
| + | |
| − | |-
| + | |
| − | |-
| + | |
| − | |-
| + | |
| − | |-
| + | |
| − | |-
| + | |
| − | |-
| + | |
| − | |-
| + | |
| − | | colspan="3" rowspan="1" | <div style="color: #000000; font-style: normal; font-weight: bold; margin-bottom: 3pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1223078"></a>Figure 61. The Image Attributes dialog box options
| + | |
| − | </div> | + | |
| | |} | | |} |
| − | </div><div style="color: #000000; font-style: normal; font-weight: bold; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 14pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1221573"></a>To modify the name of the image dataset
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220490"></a> 1 Open either a DICOM or non-DICOM image dataset.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220494"></a> 2 Select Image > Attributes > Edit Attributes in the MIPAV window. The Image Attributes dialog box opens.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220495"></a>
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220506"></a>
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220507"></a> 3 Select the name of the image in the Image name (without suffix) box.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220508"></a> 4 Type a new name of the image.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220509"></a> 5 Click either Apply or OK. The new image name appears in the header on the image window.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220514"></a>
| |
| − | {| border="1" cellpadding="5" cellspacing="0"
| |
| − | |+
| |
| − | |-
| |
| − | | <div style="color: #000000; font-style: normal; font-weight: bold; margin-bottom: 1pt; margin-left: 0pt; margin-right: 0pt; margin-top: 17pt; text-align: center; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220512"></a>What's the Difference Between
| |
| − | Clicking Apply or Clicking OK?
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 7pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220513"></a>Click Apply when you want the dialog box or window to remain open after it makes the change so that you can make further modifications. If instead you click OK, the dialog box or window closes immediately after making the current change.
| |
| − | </div>
| |
| − | |}
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: bold; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 14pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220516"></a>To edit the dimensions and units of measure
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220517"></a> 1 Open either a DICOM or non-DICOM image dataset.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220521"></a> 2 Select Image > Attributes > Edit Attributes in the MIPAV window. The Image Attributes dialog box opens.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220522"></a> 3 Click Resolutions. The Resolutions page in the Image Attributes dialog box appears.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220523"></a> 4 Type the changed values in the dimension text boxes on the left and then select the appropriate value in the Unit of measure boxes on the right.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220524"></a> 5 Click either Apply or OK.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220533"></a>
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220534"></a>
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: bold; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 14pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220536"></a>To edit the <span style="color: #000000; font-style: italic; font-weight: bold; text-decoration: none; text-transform: none; vertical-align: baseline">x, y,</span> and <span style="color: #000000; font-style: italic; font-weight: bold; text-decoration: none; text-transform: none; vertical-align: baseline">z </span>orientations of image datasets
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220537"></a> 1 Open either a DICOM or non-DICOM image dataset.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220541"></a> 2 Select Image > Attributes > Edit Attributes in the MIPAV window. The Image Attributes dialog box opens.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220545"></a> 3 Click Orientations. The Orientations page (<a href="MIPAV_ImageBasics.html#1223078">Figure 61</a>) in the Image Attributes dialog box opens.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220556"></a>
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220557"></a> 4 Select the appropriate values in the boxes on the right.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220558"></a> 5 Click Apply or OK.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: bold; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 14pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220560"></a>To change the origin for the first image slice
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220561"></a> 1 Open either a DICOM or non-DICOM image dataset.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220565"></a> 2 Select Image > Attributes > Edit Attributes in the MIPAV window. The Image Attributes dialog box opens.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220569"></a> 3 Click Orientation. The Orientation page (<a href="MIPAV_ImageBasics.html#1223078">Figure 61</a>) in the Image Attributes dialog box appears.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220579"></a>
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220580"></a> 4 Type the appropriate values in the dimension boxes.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220581"></a> 5 Click Apply or OK.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: bold; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 14pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220583"></a>To change the transform matrix
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220584"></a> 1 Open either a DICOM or non-DICOM image dataset.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220588"></a> 2 Select Image > Attributes > Edit Attributes in the MIPAV window. The Image Attributes dialog box opens.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220592"></a> 3 Click Transform Matrix. The Transform Matrix page (<a href="MIPAV_ImageBasics.html#1223078">Figure 61</a>) in the Image Attributes dialog box appears.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220593"></a>
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220594"></a>
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220595"></a>
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1220605"></a>
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220607"></a> 4 Select the appropriate transform ID in the Transform ID box.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220608"></a> 5 Load and save transform matrices.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 18pt; margin-right: 0pt; margin-top: 6pt; text-align: left; text-decoration: none; text-indent: -18pt; text-transform: none; vertical-align: baseline"><a name="1220609"></a> 6 Click Apply or OK.
| |
| − | </div><div style="color: #000000; font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><a name="1209926"></a>
| |
| − | </div><p class="Body"><a name="1191460"></a></blockquote>
| |
| − | ----
| |
| − |
| |
| − | {| align="right" border="0" cellspacing="0" cellpadding="0"
| |
| − | | align="right" | <font size="1"><a href="http://www.webworks.com"><img src="images/webworks.gif" width="150" height="20" border="0"></a>
| |
| − |
| |
| − | Quadralay Corporation
| |
| − |
| |
| − | http://www.webworks.com
| |
| − |
| |
| − | Voice: (512) 719-3399
| |
| − |
| |
| | | | |
| − | Fax: (512) 719-3606
| + | <br /> |
| − | <a href="mailto:sales@webworks.com">sales@webworks.com</a> | + | '''To edit the dimensions and units of measure'''<br /> |
| − | </font> | + | 1 Open either a DICOM or non-DICOM image dataset.<br /> |
| | + | 2 Select Image > Attributes > Edit Attributes in the MIPAV window. The Image Attributes dialog box opens.<br /> |
| | + | 3 Click Resolutions. The Resolutions page in the Image Attributes dialog box appears.<br />< |
| | + | 4 Type the changed values in the dimension text boxes on the left and then select the appropriate value in the Unit of measure boxes on the right.<br /> |
| | + | 5 Click either Apply or OK.<br /> |
| | + | '''To edit the ''x, y,'' and ''z'' orientations of image datasets<br />''' |
| | + | 1 Open either a DICOM or non-DICOM image dataset.<br /> |
| | + | 2 Select Image > Attributes > Edit Attributes in the MIPAV window. The Image Attributes dialog box opens.<br /> |
| | + | 3 Click Orientations. The Orientations page (Figure 4) in the Image Attributes dialog box opens.<br /> |
| | + | 4 Select the appropriate values in the boxes on the right.<br /> |
| | + | 5 Click Apply or OK.<br /> |
| | + | '''To change the origin for the first image slice <br />''' |
| | + | 1 Open either a DICOM or non-DICOM image dataset.<br /> |
| | + | 2 Select Image > Attributes > Edit Attributes in the MIPAV window. The Image Attributes dialog box opens.<br /> |
| | + | 3 Click Orientation. The Orientation page (Figure 4]) in the Image Attributes dialog box appears.<br /> |
| | + | 4 Type the appropriate values in the dimension boxes.<br /> |
| | + | 5 Click Apply or OK.<br /> |
| | + | '''To change the transform matrix <br />''' |
| | + | 1 Open either a DICOM or non-DICOM image dataset.<br /> |
| | + | 2 Select Image > Attributes > Edit Attributes in the MIPAV window. The Image Attributes dialog box opens.<br /> |
| | + | 3 Click Transform Matrix. The Transform Matrix page (Figure 3) in the Image Attributes dialog box appears.<br /> |
| | + | 4 Select the appropriate transform ID in the Transform ID box.<br /> |
| | + | 5 Load and save transform matrices.<br /> |
| | + | 6 Click Apply or OK.<br /> |
In this chapter. . .
This chapter also explains the dimensionality and coordinate system used by MIPAV, the data types MIPAV supports, the header offset information, endianness, and how to view and edit image attributes.It also provides a basic information about image file formats supported by MIPAV. More information about graphic file formats and the formats that MIPAV supports can be found in Appendix C: "Supported formats" .
MIPAV supports three categories of graphic file formats: vector (i.e., volume of interest, or VOI), bitmap (i.e., 2D to 5D grayscale or RGB images), and 3D images.
Vector file formats are usually used to store line-based elements. These elements can be geometric shapes, such as polygons, curves, or splines. MIPAV can read and generate several vector-formatted file types including the MIPAV lookup table (LUT), MIPAV MTX, MIPAV Graphics Plot (PLT), and MIPAV VOI. See also Appendix C: "Supported formats" .
Bitmap file formats are usually used to store real-world graphics data e.g., photographs or medical images such as CT scans. The data portion of bitmap files contains numerical data that indicates the color of each pixel or voxel in the image. MIPAV can read and generate a number of bitmap formatted file types, such as Adobe Photoshop (PSD), Graphics Interchange File (GIF), Sun Raster (RS), Tagged Image File Format (TIFF), and Truevision Graphics Adapter (TGA).
</font>MIPAV uses 3D graphic formatted files to store descriptions of the color and shape of 3D models of real-world and imaginary objects.
To support the wide range of image-processing needs of the NIH intramural research community, MIPAV reads and writes the image file types indicated in Table 1, refer to Appendix C: "Supported formats".
The voxel in the upper left-most position is considered the origin of the dataset. (A volumetric dataset is a set of cross-sectional images). Ideally, each voxel would be isotropic (i.e., cubic), but clinical datasets are often acquired in such a manner that the resolution in the x and y dimensions is greater than the resolution in the z dimension, thus producing rectangular voxels (elongated in the z. This voxel anisotropy is taken into account in the execution of the algorithms found in MIPAV.
When recorded in scanner or image order, multi-component images are stored as consecutive volumes. This is convenient for visualization, since one can easily render a particular 3D volume, however, it is inconvenient for parallel processing, as one must read the entire 4D image in order to get the specific component(s) for a particular processing.
Information about the image appears in the Image Information dialog box and in the Image Attributes dialog box.
The Image Attributes window displays the name of the image file and information about the resolution of the image. The About Image window displays more detailed information about the image, such as the orientation of the image or the number of bits per pixel or voxel.
To view the attributes of an image, select Image > Attributes > View Header in the MIPAV window. For DICOM datasets, the Image Information dialog box similar to the one shown in Figure 2-a appears. For non-DICOM datasets, an Image Information dialog box similar to the one in Figure 2-b appears.