Difference between revisions of "Image formats descriptions"
m (→Analyze files) |
m (→See also:) |
||
| (34 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | == Adobe Photoshop (PSD) files == | |
PSD is a bitmap file format created by Adobe. It can accommodate images with an unlimited amount of colors. PSD files are not multiple image files; the maximum image size is 30K x 30K pixels or voxels. PSD files are generally used to store images that were altered or manipulated by the Adobe Photoshop software. The header is embedded in the file and is 26 bytes in length. It contains information such as the height and width of the image and the color mode.<br /> | PSD is a bitmap file format created by Adobe. It can accommodate images with an unlimited amount of colors. PSD files are not multiple image files; the maximum image size is 30K x 30K pixels or voxels. PSD files are generally used to store images that were altered or manipulated by the Adobe Photoshop software. The header is embedded in the file and is 26 bytes in length. It contains information such as the height and width of the image and the color mode.<br /> | ||
<div id="AnalyzeFormat"></div> | <div id="AnalyzeFormat"></div> | ||
| − | + | == Analyze files == | |
| − | [http://en.wikipedia.org/wiki/Analyze_%28imaging_software%29 Analyze formatted files] are generated by a UNIX-based, image-processing application developed at the Mayo Clinic. | + | [http://en.wikipedia.org/wiki/Analyze_%28imaging_software%29 Analyze formatted files] are generated by a UNIX-based, image-processing application developed at the Mayo Clinic. In support of some NIH intramural researchers who have used Analyze in the past and have generated a large number of legacy datasets, MIPAV reads and writes Analyze formatted images. |
| + | |||
An image file in Analyze format contains uncompressed voxel data for the images in one of several possible voxel formats: | An image file in Analyze format contains uncompressed voxel data for the images in one of several possible voxel formats: | ||
| Line 23: | Line 24: | ||
'''Analyze images are formed from the following files:''' | '''Analyze images are formed from the following files:''' | ||
| − | '''Header file''' - This file (extension .HDR) describes the image type, size, and other important image attributes. | + | '''Header file''' - This file (extension .HDR) describes the image type, size, and other important image attributes. For example, <span style="font-family:courier">brain.hdr</span> is the header file that describes the image file. |
| − | + | ||
| − | + | ||
| − | ''' | + | '''File Containing the Actual Image Data''' - This file (extension .IMG) can be interpreted as a raw file since it does not have any header information within the file. For example, <span style="font-family:courier">brain.img</span> is the file that contains the image data. |
| − | + | '''A color lookup file''' is sometimes included in the file set - A LKUP extension indicates a lookup table file. Example: <span style="font-family:courier">file.lkup</span>. | |
| + | All files from the file set have the same name and are distinguished by the extensions HDR for the header file, IMG for image file and LKUP for lookup table file. MIPAV stores files in Analyze format in the catalog you specified in the Save Image as dialog box. | ||
| − | MIPAV could save images with *.img extension either in Analyze format or in NIFTI format. To save an image in Analyze format, call the File > Save Image as menu, and then type the file name with *.img extension. In the Choose Type of File to Write dialog box that appears, specify the file format (Analyze). Check the option Always save .img files in Analyze format, if you want MIPAV to save .img files only in that format. | + | '''Note:''' MIPAV could save images with *.img extension either in Analyze format or in [[#NiftiFormat| NIFTI format]]. To save an image in Analyze format, call the File > Save Image as menu, and then type the file name with *.img extension. In the Choose Type of File to Write dialog box that appears, specify the file format (Analyze). Check the option Always save .img files in Analyze format, if you want MIPAV to save .img files only in that format. |
| − | + | For more information about Analyze file format, please refer to the [http://web.archive.org/web/20070927191351/http://www.mayo.edu/bir/PDF/ANALYZE75.pdf| ANALYZE 7.5. format] document from [http://mayoresearch.mayo.edu/mayo/research/robb_lab/analyze.cfm the Mayo Clinic]. | |
| − | + | == AFNI == | |
<span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline">'''<font color="#000000">The HEAD file</font>'''</span> for an AFNI dataset is in ASCII, so you can view it with a normal text editor (e.g., Notepad). The data within are organized into <span style="font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">''<font color="#000000">attributes</font>''</span>, which are named arrays of floats, integers, or characters (strings). A sample float attribute is shown in Table 2. This array defines the voxel array dimensions. An example of a character array attribute is shown in Table 3. | <span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline">'''<font color="#000000">The HEAD file</font>'''</span> for an AFNI dataset is in ASCII, so you can view it with a normal text editor (e.g., Notepad). The data within are organized into <span style="font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">''<font color="#000000">attributes</font>''</span>, which are named arrays of floats, integers, or characters (strings). A sample float attribute is shown in Table 2. This array defines the voxel array dimensions. An example of a character array attribute is shown in Table 3. | ||
| Line 70: | Line 70: | ||
<div id="AviFormat"></div> | <div id="AviFormat"></div> | ||
| − | + | == Audio Video Interleave (AVI) files == | |
<div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> AVI is the Microsoft Video for Windows standard. AVI is a form of the Resource Interchange File Format (RIFF). In this file format, video and audio data are stored consecutively in an AVI file. The AVI file contains a 4-byte file header, followed by list information and then alternating video and audio streams.<br /></font></div> | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> AVI is the Microsoft Video for Windows standard. AVI is a form of the Resource Interchange File Format (RIFF). In this file format, video and audio data are stored consecutively in an AVI file. The AVI file contains a 4-byte file header, followed by list information and then alternating video and audio streams.<br /></font></div> | ||
| − | + | == BIORAD == | |
<div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> [http://www.aecom.yu.edu/aif/instructions/BioRad_instructions/formattradiance.htm Each Biorad confocal image file consists of three parts. They are 1) a 76 byte header which contains information such as the number of images in the file and how large each section is, 2) the images themselves and 3) notes after the images. Each set of BIORAD images from one set of raw data, is stored in a separate folder with the name provided. Within that folder is a subfolder named /]<span style="font-style: normal; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline"><font color="#000000">Raw Data/</font></span> where the files are stored. A single image file is called <span style="font-style: normal; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline"><font color="#000000">raw.pic</font></span>. In the multiple probe Z or T series, the files are named <span style="font-style: normal; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline"><font color="#000000">raw01.pic</font></span>, <span style="font-style: normal; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline"><font color="#000000">raw02.pic</font></span>, etc. There are may be also text files stored in the same folder, which contain information about the imaging parameters used.<br /></font></div> | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> [http://www.aecom.yu.edu/aif/instructions/BioRad_instructions/formattradiance.htm Each Biorad confocal image file consists of three parts. They are 1) a 76 byte header which contains information such as the number of images in the file and how large each section is, 2) the images themselves and 3) notes after the images. Each set of BIORAD images from one set of raw data, is stored in a separate folder with the name provided. Within that folder is a subfolder named /]<span style="font-style: normal; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline"><font color="#000000">Raw Data/</font></span> where the files are stored. A single image file is called <span style="font-style: normal; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline"><font color="#000000">raw.pic</font></span>. In the multiple probe Z or T series, the files are named <span style="font-style: normal; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline"><font color="#000000">raw01.pic</font></span>, <span style="font-style: normal; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline"><font color="#000000">raw02.pic</font></span>, etc. There are may be also text files stored in the same folder, which contain information about the imaging parameters used.<br /></font></div> | ||
<div id="BrukerFormat"></div> | <div id="BrukerFormat"></div> | ||
| − | + | == Bruker data format == | |
Bruker format stores a single scanning session in its own directory. The directory is named according to the subject name or number, as typed in by the scanner operator. The directory name usually specifies the subject name/number and which session this is for that subject. For more information, refer to <<span style="font-style: normal; font-weight: normal; text-transform: none; vertical-align: baseline"><u><font color="#000000">http://imaging.mrc-cbu.cam.ac.uk/imaging/FormatBruker</font></u></span>>. | Bruker format stores a single scanning session in its own directory. The directory is named according to the subject name or number, as typed in by the scanner operator. The directory name usually specifies the subject name/number and which session this is for that subject. For more information, refer to <<span style="font-style: normal; font-weight: normal; text-transform: none; vertical-align: baseline"><u><font color="#000000">http://imaging.mrc-cbu.cam.ac.uk/imaging/FormatBruker</font></u></span>>. | ||
| − | + | == Cheschire (IMG or IMC) == | |
<div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> TBD.<br /></font></div> | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> TBD.<br /></font></div> | ||
<div id="DicomFormat"></div> | <div id="DicomFormat"></div> | ||
| − | + | == Digital Imaging and Communications in Medicine (DICOM) files == | |
The DICOM format is a standard that defines a standard method of communication between two devices, such as computers, servers, or imaging devices. DICOM not only prescribes how to communicate with other imaging equipment and databases but also specifies how images are stored. Each image has accompanying header information describing the image format (i.e., height, width, etc.) as well as information that indirectly relates to the image (i.e., patient information, image equipment setup parameters). | The DICOM format is a standard that defines a standard method of communication between two devices, such as computers, servers, or imaging devices. DICOM not only prescribes how to communicate with other imaging equipment and databases but also specifies how images are stored. Each image has accompanying header information describing the image format (i.e., height, width, etc.) as well as information that indirectly relates to the image (i.e., patient information, image equipment setup parameters). | ||
| Line 97: | Line 97: | ||
<div id="FitsFormat"></div> | <div id="FitsFormat"></div> | ||
| − | + | == FITS == | |
FITS stands for "Flexible Image Transport System" and is the standard astronomical data format endorsed by both NASA and the IAU. FITS is primarily designed to store scientific data sets consisting of multi-dimensional arrays (1-D spectra, 2-D images or 3-D data cubes) and 2-dimensional tables containing rows and columns of data. | FITS stands for "Flexible Image Transport System" and is the standard astronomical data format endorsed by both NASA and the IAU. FITS is primarily designed to store scientific data sets consisting of multi-dimensional arrays (1-D spectra, 2-D images or 3-D data cubes) and 2-dimensional tables containing rows and columns of data. | ||
| Line 105: | Line 105: | ||
<div id="FreeSurferFormat"></div> | <div id="FreeSurferFormat"></div> | ||
| − | + | == FreeSurfer image and surface files == | |
FreeSurfer uses two main kinds of data. The first one is a volume of voxels, as for example, from an MRI scanner. It provides a source of raw input. The other one is surface data which consist of lists of vertices as well as their positions in space and faces and also vertices associated with them. | FreeSurfer uses two main kinds of data. The first one is a volume of voxels, as for example, from an MRI scanner. It provides a source of raw input. The other one is surface data which consist of lists of vertices as well as their positions in space and faces and also vertices associated with them. | ||
| Line 114: | Line 114: | ||
<div id="GenesisFormat"></div> | <div id="GenesisFormat"></div> | ||
| − | + | ==GE - Genesis 5X and LX== | |
The format of the headers is General Electric Genesis described in the [[http://www.dclunie.com/medical-image-faq/html/part8.html| Medical Image Format FAQ]]. The image header contains the table position and field of view. This is important information, because the spacing between the slices and the pixel size changes several times throughout the data set. | The format of the headers is General Electric Genesis described in the [[http://www.dclunie.com/medical-image-faq/html/part8.html| Medical Image Format FAQ]]. The image header contains the table position and field of view. This is important information, because the spacing between the slices and the pixel size changes several times throughout the data set. | ||
| Line 126: | Line 126: | ||
*http://www.genesismedicalimaging.com/mobiles/mobilemri.html | *http://www.genesismedicalimaging.com/mobiles/mobilemri.html | ||
| − | + | == Graphics Interchange Format (GIF) files == | |
| − | [http://en.wikipedia.org/wiki/GIF The Graphics Interchange Format (GIF) is a bitmap file format that was created by CompuServe, Inc. GIF is primarily an exchange and storage file format; GIFs can be used to store one or several bitmap images in one file. GIFs support pixel or voxel depths of 1 to 8 bits. The maximum image size is 64K x 64K pixel or voxels. There are two major revisions of the GIF format specification: GIF87a and GIF89a. Both formats begin with a 6-byte header that identifies the file format as GIF. | + | [http://en.wikipedia.org/wiki/GIF The Graphics Interchange Format (GIF)] is a bitmap file format that was created by CompuServe, Inc. GIF is primarily an exchange and storage file format; GIFs can be used to store one or several bitmap images in one file. GIFs support pixel or voxel depths of 1 to 8 bits. The maximum image size is 64K x 64K pixel or voxels. There are two major revisions of the GIF format specification: GIF87a and GIF89a. Both formats begin with a 6-byte header that identifies the file format as GIF.<br /> |
<div id="ImageCytometryFormat"></div> | <div id="ImageCytometryFormat"></div> | ||
| − | + | == ICS (Image Cytometry Standard) == | |
The Image Cytometry Standard (ICS) is a digital multidimensional image file format used in life sciences microscopy. It stores the image data and the microscopic parameters describing the optics during the acquisition. The newest ICS2 file format uses a single <span style="font-style: normal; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline"><font color="#000000">ICS</font></span> file with both the header and the data together. | The Image Cytometry Standard (ICS) is a digital multidimensional image file format used in life sciences microscopy. It stores the image data and the microscopic parameters describing the optics during the acquisition. The newest ICS2 file format uses a single <span style="font-style: normal; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline"><font color="#000000">ICS</font></span> file with both the header and the data together. | ||
| Line 139: | Line 139: | ||
For more information, refer to http://en.wikipedia.org/wiki/Image_Cytometry_Standard. | For more information, refer to http://en.wikipedia.org/wiki/Image_Cytometry_Standard. | ||
| − | + | == Joint Photographic Experts Group (JPEG/JFIF) files == | |
<div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> [http://en.wikipedia.org/wiki/JPEG The JPEG File Interchange Format (JFIF) allows files containing JPEG-encoded data streams to be exchanged. Technically, JPEG refers to the compression type and the Joint Photographic Experts Group standards organization. However, the term ]<span style="font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">''<font color="#000000">JPEG</font>''</span> is usually used to indicate the file format. JPEG file formats are bitmap files that are primarily used in image and graphics manipulation programs. Created by C-Cube Microsystems, the JPEG or JFIF file format does not accommodate multiple images per file and the maximum image size allowed is 64K by 64K pixel or voxels. JPEG files can accommodate 24-bit color images. Generally, JPEG header information appears between the Start of Image (SOI) and Application (APP0) markers.<br /> | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> [http://en.wikipedia.org/wiki/JPEG The JPEG File Interchange Format (JFIF) allows files containing JPEG-encoded data streams to be exchanged. Technically, JPEG refers to the compression type and the Joint Photographic Experts Group standards organization. However, the term ]<span style="font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">''<font color="#000000">JPEG</font>''</span> is usually used to indicate the file format. JPEG file formats are bitmap files that are primarily used in image and graphics manipulation programs. Created by C-Cube Microsystems, the JPEG or JFIF file format does not accommodate multiple images per file and the maximum image size allowed is 64K by 64K pixel or voxels. JPEG files can accommodate 24-bit color images. Generally, JPEG header information appears between the Start of Image (SOI) and Application (APP0) markers.<br /> | ||
| + | <div id="LaunchAnywhereFiles"></div> | ||
<div id="ZeissFormat"></div> | <div id="ZeissFormat"></div> | ||
| − | + | ||
| + | == Laser Scanning Microscope (Zeiss) == | ||
Laser Scanning Microscope (LSM) is a line of laser point scanning microscopes produced by [http://www.zeiss.de/lsm700 Carl Zeiss LLC]. The new LSM 700/710/780 series were released after 2008, thus making the 5th generation, including the LSM 510, LSM 5 Pascal, and LSM 5 Live obsolete. | Laser Scanning Microscope (LSM) is a line of laser point scanning microscopes produced by [http://www.zeiss.de/lsm700 Carl Zeiss LLC]. The new LSM 700/710/780 series were released after 2008, thus making the 5th generation, including the LSM 510, LSM 5 Pascal, and LSM 5 Live obsolete. | ||
| Line 153: | Line 155: | ||
<div id="LegacyVtkFormat"></div> | <div id="LegacyVtkFormat"></div> | ||
| − | + | == Legacy VTK == | |
The legacy VTK file format is a surface format that consist of five basic parts: | The legacy VTK file format is a surface format that consist of five basic parts: | ||
| Line 167: | Line 169: | ||
<div id="LiffFormat"></div> | <div id="LiffFormat"></div> | ||
| − | + | == LIFF == | |
Layered Image File Format is a file format used in the [http://www.perkinelmer.com/pages/020/cellularimaging/products/openlab.xhtml Openlab] suite for microscope image processing. It is a proprietary format, but has an open, extensible form analogous to TIFF. It was specifically designed to contain a large number of high resolution images, and also all of the meta data generated by analysis of such images. | Layered Image File Format is a file format used in the [http://www.perkinelmer.com/pages/020/cellularimaging/products/openlab.xhtml Openlab] suite for microscope image processing. It is a proprietary format, but has an open, extensible form analogous to TIFF. It was specifically designed to contain a large number of high resolution images, and also all of the meta data generated by analysis of such images. | ||
| Line 173: | Line 175: | ||
For more information, refer to [http://www.perkinelmer.com/pages/020/cellularimaging/products/openlab.xhtml OpenLab web site]. | For more information, refer to [http://www.perkinelmer.com/pages/020/cellularimaging/products/openlab.xhtml OpenLab web site]. | ||
| − | + | == Macintosh PICT == | |
[http://www.fileformat.info/format/macpict/egff.htm The Macintosh PICT (Macintosh Picture) format is format for the Macintosh. It also known as PICT, Macintosh Picture, or QuickDraw Picture Format. All Macintosh PICT files start with a 512-byte header, which contains information that the Macintosh uses to keep track of the file. This is followed by three fields describing the image size (picSize), the image frame (picFrame), and a version number. For more information, refer to the Macintosh PICT File Format Summary page at <]<span style="font-style: normal; font-weight: normal; text-transform: none; vertical-align: baseline"><u><font color="#000000">http://www.fileformat.info/format/macpict/egff.htm</font></u></span>>.<br /> | [http://www.fileformat.info/format/macpict/egff.htm The Macintosh PICT (Macintosh Picture) format is format for the Macintosh. It also known as PICT, Macintosh Picture, or QuickDraw Picture Format. All Macintosh PICT files start with a 512-byte header, which contains information that the Macintosh uses to keep track of the file. This is followed by three fields describing the image size (picSize), the image frame (picFrame), and a version number. For more information, refer to the Macintosh PICT File Format Summary page at <]<span style="font-style: normal; font-weight: normal; text-transform: none; vertical-align: baseline"><u><font color="#000000">http://www.fileformat.info/format/macpict/egff.htm</font></u></span>>.<br /> | ||
<div id="MincFormat"></div> | <div id="MincFormat"></div> | ||
| − | + | == Medical Image NetCDF (MINC) files == | |
[http://www.bic.mni.mcgill.ca/software/minc/ MINC] is a medical-imaging file format that is based on the Network Common Data Form (NetCDF) file format. NetCDF is a platform-independent software interface that provides a means for storing named, multidimensional variables. Each multidimensional variable is defined by a name, dimensions, and attributes. For example, cardio (name) [256, 256] (dimensions), and "long_name" where this attribute is a string that describes the content of the image. MINC provides a standard for dimension, variable, and attribute names suitable for medical imaging. MINC also provides convenience functions to complement the NetCDF interface and convenience functions for using MINC files. | [http://www.bic.mni.mcgill.ca/software/minc/ MINC] is a medical-imaging file format that is based on the Network Common Data Form (NetCDF) file format. NetCDF is a platform-independent software interface that provides a means for storing named, multidimensional variables. Each multidimensional variable is defined by a name, dimensions, and attributes. For example, cardio (name) [256, 256] (dimensions), and "long_name" where this attribute is a string that describes the content of the image. MINC provides a standard for dimension, variable, and attribute names suitable for medical imaging. MINC also provides convenience functions to complement the NetCDF interface and convenience functions for using MINC files. | ||
| Line 184: | Line 186: | ||
<div id="MrcFormat"></div> | <div id="MrcFormat"></div> | ||
| − | + | == Medical Research Council (MRC) == | |
The MRC file format is a file format for electron density. It is an industry standard for Cryo-electron microscopy where the result of the technique is a three-dimensional grid of voxels each with a value corresponding to density of electrons. The MRC format is supported by almost every molecular graphics suite that supports volumetric data. | The MRC file format is a file format for electron density. It is an industry standard for Cryo-electron microscopy where the result of the technique is a three-dimensional grid of voxels each with a value corresponding to density of electrons. The MRC format is supported by almost every molecular graphics suite that supports volumetric data. | ||
| Line 191: | Line 193: | ||
<div id="MicroCat"></div> | <div id="MicroCat"></div> | ||
| − | + | == MICRO-CAT == | |
Siemens microCAT image format is designed for its microCAT+SPECT high resolution, high sensitivity, in vivo | Siemens microCAT image format is designed for its microCAT+SPECT high resolution, high sensitivity, in vivo | ||
| Line 198: | Line 200: | ||
For more information refer to: [http://www.medical.siemens.com/siemens/en_US/rg_marcom_FBAs/files/Press_Releases/Siemens_Says/JuneSiemensSays.pdf Siemens press release.] | For more information refer to: [http://www.medical.siemens.com/siemens/en_US/rg_marcom_FBAs/files/Press_Releases/Siemens_Says/JuneSiemensSays.pdf Siemens press release.] | ||
| − | + | == Microsoft Windows Bitmap (BMP) files == | |
| − | + | [http://en.wikipedia.org/wiki/Windows_and_OS/2_bitmap BMP] is a native bitmap format for the Microsoft Windows and OS/2 platforms. Developed by the Microsoft Corporation, it can be read on Intel machines running Microsoft Windows, Windows XP, Windows NT, Windows 95, OS/2, and MS-DOS. It is an uncompressed file format with a maximum image size of 32K x 32K or 2G x 2G pixels or voxels (depending on the version of BMP).br /> | |
Depending on the version of BMP, for OS/2 the maximum image size is larger, 64K x 64K or 4G x 4G pixels or voxels. There are several versions of BMP for Microsoft Windows and OS/2. MIPAV supports Microsoft Window BMP versions 2.x and above and all OS/2 versions of the BMP file format. BMP version 2.x is designed for use with the Microsoft Windows 2.x platform. It has a 14-byte header (as does the OS/2 1.x bitmap header). It can accommodate images with 1-, 4-, 8-, or 24-bit colors. Version 3.x is designed for use with the Microsoft Windows 3.x and Windows NT platforms. Like version 2.x, it contains a 14-byte header. It also contains an additional bitmap header that is 40 bytes in size. The Microsoft Windows 3.x platform version accommodates images with 1-, 4-, 8- or 24-bit colors. Windows NT and Windows XP accommodate 16- and 32-bit images. Version 4.x was designed for use with Microsoft Windows 95. It contains the 14-byte header, and an additional 108-byte bitmap header. It can accommodate images of 1, 4, 8, 16, and 32 bits. | Depending on the version of BMP, for OS/2 the maximum image size is larger, 64K x 64K or 4G x 4G pixels or voxels. There are several versions of BMP for Microsoft Windows and OS/2. MIPAV supports Microsoft Window BMP versions 2.x and above and all OS/2 versions of the BMP file format. BMP version 2.x is designed for use with the Microsoft Windows 2.x platform. It has a 14-byte header (as does the OS/2 1.x bitmap header). It can accommodate images with 1-, 4-, 8-, or 24-bit colors. Version 3.x is designed for use with the Microsoft Windows 3.x and Windows NT platforms. Like version 2.x, it contains a 14-byte header. It also contains an additional bitmap header that is 40 bytes in size. The Microsoft Windows 3.x platform version accommodates images with 1-, 4-, 8- or 24-bit colors. Windows NT and Windows XP accommodate 16- and 32-bit images. Version 4.x was designed for use with Microsoft Windows 95. It contains the 14-byte header, and an additional 108-byte bitmap header. It can accommodate images of 1, 4, 8, 16, and 32 bits. | ||
<div id="MipavLutFormat"></div> | <div id="MipavLutFormat"></div> | ||
| − | + | == MIPAV (LUT) files == | |
LUT is a vector file format used to store MIPAV Lookup Table ( [[Visualizing Images: Displaying images using the default view:Changing image brightness and contrast using LUTs#WhatIsLut | LUT]]) data. | LUT is a vector file format used to store MIPAV Lookup Table ( [[Visualizing Images: Displaying images using the default view:Changing image brightness and contrast using LUTs#WhatIsLut | LUT]]) data. | ||
| Line 211: | Line 213: | ||
MIPAV provides a logical color map, which allows you to remap the original intensities to other intensities. Although technically the term ''look-up table'' (LUT) can be used for the physical and logical color maps, in this guide look-up table refers to the logical color map only. You can apply predefined, pseudo color or inverse LUTs, or you can manually manipulate the transfer function used to map the image data to the LUT. The LUT then translates the remapped values so that they can be interpreted by the physical color map and displayed on your monitor. | MIPAV provides a logical color map, which allows you to remap the original intensities to other intensities. Although technically the term ''look-up table'' (LUT) can be used for the physical and logical color maps, in this guide look-up table refers to the logical color map only. You can apply predefined, pseudo color or inverse LUTs, or you can manually manipulate the transfer function used to map the image data to the LUT. The LUT then translates the remapped values so that they can be interpreted by the physical color map and displayed on your monitor. | ||
| − | For a sample LUT file refer to [[ | + | For a sample LUT file refer to [[Matrix, Plot, LUT, VOI and Surface files#MipavLutFile| LUT file]].<br /> |
<div id="MipavMtl"></div> | <div id="MipavMtl"></div> | ||
| − | + | ||
| + | == MIPAV (MTX) files == | ||
The matrix filedescribes the image's orientation, translations, offset scales, rotations, and shears. When any of this information changes in the image, the identity matrix changes. | The matrix filedescribes the image's orientation, translations, offset scales, rotations, and shears. When any of this information changes in the image, the identity matrix changes. | ||
| Line 221: | Line 224: | ||
<div id="MipavPlotFiles"></div> | <div id="MipavPlotFiles"></div> | ||
| − | + | == MIPAV (PLT) files == | |
The MIPAV plot file (.PLT) is a vector file that is used to store graphics data. For a sample PLT file refer to [[Matrix, Plot, VOI and Surface files#PlotFile| Plot file]].<br /> | The MIPAV plot file (.PLT) is a vector file that is used to store graphics data. For a sample PLT file refer to [[Matrix, Plot, VOI and Surface files#PlotFile| Plot file]].<br /> | ||
| − | + | == MIPAV text file format == | |
| − | MIPAV defined ASCII text surface format. The text surface file records the triangle mesh vertices, normals and connectivity. | + | MIPAV defined ASCII text surface format. The text surface file records the triangle mesh vertices, normals and connectivity. |
| + | |||
| + | <div style="font-family:courier;font-size:-1"> | ||
| + | Vertices | ||
| + | x, y, z // vertex position | ||
| + | |||
| + | Normal | ||
| + | |||
| + | x, y, z // normal coordinate | ||
| + | |||
| + | Connectivity | ||
| + | |||
| + | x, y, z // index connection | ||
| + | </div> | ||
| + | |||
| + | See also: [[Matrix, Plot, VOI and Surface files#MipavTextFormat| MIPAV surface file in text format]]. <br /> | ||
<div id="MipavSurfaceFormat"></div> | <div id="MipavSurfaceFormat"></div> | ||
| − | + | ||
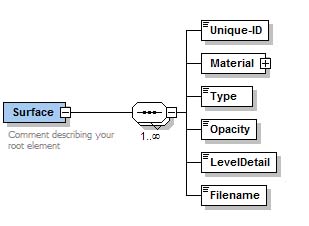
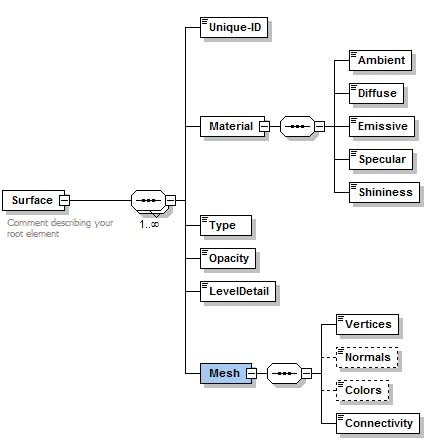
| + | == MIPAV (SUR) files == | ||
MIPAV supports its own surface format (*.sur). A MIPAV surface file contains the information about the surface area and geometry, plus the additional information that describes the surface color and more. This is defined by [http://mipav.cit.nih.gov/documentation/xml-format/image/ an XML scheme], which can be found on [http://mipav.cit.nih.gov/documentation/xml-format/image/ the MIPAV web site]. | MIPAV supports its own surface format (*.sur). A MIPAV surface file contains the information about the surface area and geometry, plus the additional information that describes the surface color and more. This is defined by [http://mipav.cit.nih.gov/documentation/xml-format/image/ an XML scheme], which can be found on [http://mipav.cit.nih.gov/documentation/xml-format/image/ the MIPAV web site]. | ||
| Line 261: | Line 280: | ||
See also [[Matrix, Plot, VOI and Surface files#MipavSurfaceFiles| MIPAV Surface files description]]. | See also [[Matrix, Plot, VOI and Surface files#MipavSurfaceFiles| MIPAV Surface files description]]. | ||
| − | + | == MIPAV (VOI) files == | |
VOI is a vector file format that is used to store volume of interest contouring information. For a sample file refer to [MIPAV_AppCSupportedFormats.html#1249118 "VOI file" on page 603].<br /> | VOI is a vector file format that is used to store volume of interest contouring information. For a sample file refer to [MIPAV_AppCSupportedFormats.html#1249118 "VOI file" on page 603].<br /> | ||
| − | + | == MIPAV XML == | |
| − | Refer to | + | Refer to [[Supported Formats#MIPAV XML Format| MIPAV XML]]. |
| − | + | MIPAV image schema definition [http://mipav.cit.nih.gov/documentation/xml-format/image/] | |
| − | + | == MIPAV XML surface format == | |
| − | + | See also [[Supported Formats#MipavImageSchema|MIPAV Image Schema]]. | |
| − | <div | + | MIPAV Image Schema can also be found [http://mipav.cit.nih.gov/documentation/xml-format/image/image.xsd here]. |
| + | |||
| + | <div id="NiftiFormat"></div> | ||
| + | |||
| + | == NIFTI == | ||
| + | |||
| + | [http://www.mayo.edu/bir/PDF/ANALYZE75.pdf NifTy-1] is a new Analyze-style data format, proposed by the NIfTI DFWG. NIfTI-1 was adapted from ANALYZE 7.5 file format. See also [http://nifti.nimh.nih.gov/ NIFTI]. | ||
| + | |||
| + | It uses the "empty space" in the ANALYZE 7.5 header to add several new features, which are listed below:<br / | ||
| + | ></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 36pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: -12pt; text-transform: none; vertical-align: baseline"><font color="#000000"> Affine coordinate definitions relating voxel index (i,j,k) to spatial location (x,y,z);<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 36pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: -12pt; text-transform: none; vertical-align: baseline"><font color="#000000"> Codes to indicate spatio-temporal slice ordering for FMRI;<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 36pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: -12pt; text-transform: none; vertical-align: baseline"><font color="#000000"> "Complete" set of 8-128 bit data types;<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 36pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: -12pt; text-transform: none; vertical-align: baseline"><font color="#000000"> Standardized way to store vector-valued datasets over 1-4 dimensional domains;<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 36pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: -12pt; text-transform: none; vertical-align: baseline"><font color="#000000"> Codes to indicate data "meaning";<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 36pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: -12pt; text-transform: none; vertical-align: baseline"><font color="#000000"> A standardized way to add "extension" data to the header;<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 36pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: -12pt; text-transform: none; vertical-align: baseline"><font color="#000000"> Dual file (<span style="font-style: normal; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline"><font color="#000000">hdr </font></span>& <span style="font-style: normal; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline"><font color="#000000">img</font></span>) or single file (<span style="font-style: normal; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline"><font color="#000000">nii</font></span>) storage; <br /></font></div> | ||
For more information, refer to the NIfTI web site at <<span style="font-style: normal; font-weight: normal; text-transform: none; vertical-align: baseline"><u><font color="#000000">http://nifti.nimh.nih.gov/</font></u></span>>. | For more information, refer to the NIfTI web site at <<span style="font-style: normal; font-weight: normal; text-transform: none; vertical-align: baseline"><u><font color="#000000">http://nifti.nimh.nih.gov/</font></u></span>>. | ||
| − | = | + | <div id="PaintbrushFiles"></div> |
| + | |||
| + | == PC Paintbrush (PCX) files == | ||
PC Paintbrush is a bitmap file format that is used primarily in Microsoft Windows and other Windows-based products. PCX is mainly used as an exchange and storage format. Created by ZSoft and packaged with Microsoft Windows, PCX can accommodate 1-, 4-, 8-, and 24-bit color images. It is uncompressed; the maximum image size is 64K x 64K pixel/voxels. Header information is embedded in the file; the first 128 pixel/voxels in the file contain information such as the PCX ID number, the bits per pixel/voxel, and the palette type.<br /> | PC Paintbrush is a bitmap file format that is used primarily in Microsoft Windows and other Windows-based products. PCX is mainly used as an exchange and storage format. Created by ZSoft and packaged with Microsoft Windows, PCX can accommodate 1-, 4-, 8-, and 24-bit color images. It is uncompressed; the maximum image size is 64K x 64K pixel/voxels. Header information is embedded in the file; the first 128 pixel/voxels in the file contain information such as the PCX ID number, the bits per pixel/voxel, and the palette type.<br /> | ||
| − | = | + | <div id="PictFormat"></div> |
| − | See also | + | == PICT files == |
| + | See also [[Supported Formats#PictFormat|Supported file formats - PICT]]. | ||
| − | + | == Phillips PAR/REC == | |
| − | [http://www.dclunie.com/medical-image-faq/html/part8.html See also <http://www.dclunie.com/medical-image-faq/html/part8.html>]<br / | + | [http://www.dclunie.com/medical-image-faq/html/part8.html See also <http://www.dclunie.com/medical-image-faq/html/part8.html>]<br /> |
and MIPAV User Guide, Volume 2, Algorithms "DTI Create List File". | and MIPAV User Guide, Volume 2, Algorithms "DTI Create List File". | ||
<div id="PolyFormat"></div> | <div id="PolyFormat"></div> | ||
| − | + | ||
| + | == POLY == | ||
<div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> The common <span style="font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">''<font color="#000000">poly</font>''</span> file is represented by a printable ASCII file. It contains a number of features combined in three sections: a vertex list, a polygon list, and a surface list. <br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> Each of lists is represented by a sequence of non-empty lines, called a section. The first line of a section is called its title.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> The title of a section contains its name optionally followed by the attribute list enclosed in parentheses. Several attributes in the list are separated by . Both the name and the attributes are built only of alphanumeric characters (letters, digits, underscore). By convention, data section names are written in upper case. <br /></font></div> | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> The common <span style="font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">''<font color="#000000">poly</font>''</span> file is represented by a printable ASCII file. It contains a number of features combined in three sections: a vertex list, a polygon list, and a surface list. <br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> Each of lists is represented by a sequence of non-empty lines, called a section. The first line of a section is called its title.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> The title of a section contains its name optionally followed by the attribute list enclosed in parentheses. Several attributes in the list are separated by . Both the name and the attributes are built only of alphanumeric characters (letters, digits, underscore). By convention, data section names are written in upper case. <br /></font></div> | ||
| Line 319: | Line 351: | ||
<div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> `surf %d: [%d] %d'<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> The first number is the index of the surface (numbered from zero). The second number [inside square brackets] is the number of polygons belonging to that surface. The third number is the index of <span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline">'''<font color="#000000">the first polygon in the group</font>'''</span>.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> The polygons grouped into a surface must occupy a contiguous sub-list of the polygon list. <br /></font></div> | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> `surf %d: [%d] %d'<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> The first number is the index of the surface (numbered from zero). The second number [inside square brackets] is the number of polygons belonging to that surface. The third number is the index of <span style="font-style: normal; text-decoration: none; text-transform: none; vertical-align: baseline">'''<font color="#000000">the first polygon in the group</font>'''</span>.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> The polygons grouped into a surface must occupy a contiguous sub-list of the polygon list. <br /></font></div> | ||
| − | + | == Polygon File Format == | |
<div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> Also known as Stanford Triangle Format. <br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> File Structure <br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><font color="#000000"> Header<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><font color="#000000"> Vertex List<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><font color="#000000"> Face List<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><font color="#000000"> (lists of other elements)<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> File Format ( ASCII description )<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> Also known as Stanford Triangle Format. <br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> File Structure <br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><font color="#000000"> Header<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><font color="#000000"> Vertex List<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><font color="#000000"> Face List<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><font color="#000000"> (lists of other elements)<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 24pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: -24pt; text-transform: none; vertical-align: baseline"><font color="#000000"> <br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> File Format ( ASCII description )<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> | ||
| Line 332: | Line 364: | ||
<br /></font></div> | <br /></font></div> | ||
| − | + | == Portable Network Graphic Format (PNG) files == | |
<div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> PNG is a bitmap file format that is generally used to transmit and store network image data. The PNG format can store images with up to 16-bits (grayscale) or 48-bits (truecolor) per pixel/voxel. The maximum image size is 2G x 2G pixel/voxels. PNG is a compressed format. Multiple images cannot be stored in PNG. An 8-byte identification signature is followed by a header chunk, which contains basic information about the image data. The header chunk data area is 13 bytes in length.<br /></font></div> | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> PNG is a bitmap file format that is generally used to transmit and store network image data. The PNG format can store images with up to 16-bits (grayscale) or 48-bits (truecolor) per pixel/voxel. The maximum image size is 2G x 2G pixel/voxels. PNG is a compressed format. Multiple images cannot be stored in PNG. An 8-byte identification signature is followed by a header chunk, which contains basic information about the image data. The header chunk data area is 13 bytes in length.<br /></font></div> | ||
| − | == | + | == QuickTime-Apple == |
<div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> The QuickTime file is a container file that contains one or more tracks, where track stores a particular type of data e.g., audio, video, effects, or text. Each track either contains a digitally-encoded media stream (using a specific codec) or a data reference to the media stream located in another file. Tracks are maintained in a hierarchal data structure consisting of objects called <span style="font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">''<font color="#000000">atoms</font>''</span>. An atom can be a parent to other atoms or it can contain media or edit data, but it cannot do both.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> [http://en.wikipedia.org/wiki/QuickTime#QuickTime_file_format See also: <]<span style="font-style: normal; font-weight: normal; text-transform: none; vertical-align: baseline"><u><font color="#000000">http://en.wikipedia.org/wiki/QuickTime#QuickTime_file_format</font></u></span>><br /></font></div> | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> The QuickTime file is a container file that contains one or more tracks, where track stores a particular type of data e.g., audio, video, effects, or text. Each track either contains a digitally-encoded media stream (using a specific codec) or a data reference to the media stream located in another file. Tracks are maintained in a hierarchal data structure consisting of objects called <span style="font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline">''<font color="#000000">atoms</font>''</span>. An atom can be a parent to other atoms or it can contain media or edit data, but it cannot do both.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 5pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> [http://en.wikipedia.org/wiki/QuickTime#QuickTime_file_format See also: <]<span style="font-style: normal; font-weight: normal; text-transform: none; vertical-align: baseline"><u><font color="#000000">http://en.wikipedia.org/wiki/QuickTime#QuickTime_file_format</font></u></span>><br /></font></div> | ||
| − | + | See also: [http://en.wikipedia.org/wiki/QuickTime QuickTime-Apple]. | |
| + | |||
| + | == Raw data files == | ||
<div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> MIPAV supports reading and writing Raw image data of all the basic programming types (i.e., boolean, byte, short, etc.). Raw images have no header or a header of known length and unknown content at a fixed location at the beginning of the file. You must specify basic information about the Raw image before it is loaded in MIPAV. When loading a RAW data set, you must specify the image type, dimension and resolution, units, and header offset.<br /></font></div> | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> MIPAV supports reading and writing Raw image data of all the basic programming types (i.e., boolean, byte, short, etc.). Raw images have no header or a header of known length and unknown content at a fixed location at the beginning of the file. You must specify basic information about the Raw image before it is loaded in MIPAV. When loading a RAW data set, you must specify the image type, dimension and resolution, units, and header offset.<br /></font></div> | ||
| − | == | + | == Siemens - Magnetom Vision == |
| − | Refer to | + | Refer to http://www.dclunie.com/medical-image-faq/html/part4.html#MagnetomVision and [http://www.faqs.org/faqs/medical-image-faq/part4/ Siemens - Magnetom Vision]. |
| − | + | == STL (ASCII and Binary) == | |
<div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> [http://en.wikipedia.org/wiki/3D_Systems STL is a file format initially designed for the CAD software created by 3D Systems. The STL format specifies both ASCII and binary representations. An STL file describes an unstructured triangulated surface by the unit normal and vertices, which are ordered by the right-hand rule, using a three-dimensional Cartesian coordinate system. It describes only the surface geometry of a three dimensional object without any representation of color, texture or attributes. ]<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> [http://rpdrc.ic.polyu.edu.hk/old_files/stl_ascii_format.htm See also <http://rpdrc.ic.polyu.edu.hk/old_files/stl_ascii_format.htm>.]<br /></font></div> | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> [http://en.wikipedia.org/wiki/3D_Systems STL is a file format initially designed for the CAD software created by 3D Systems. The STL format specifies both ASCII and binary representations. An STL file describes an unstructured triangulated surface by the unit normal and vertices, which are ordered by the right-hand rule, using a three-dimensional Cartesian coordinate system. It describes only the surface geometry of a three dimensional object without any representation of color, texture or attributes. ]<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> [http://rpdrc.ic.polyu.edu.hk/old_files/stl_ascii_format.htm See also <http://rpdrc.ic.polyu.edu.hk/old_files/stl_ascii_format.htm>.]<br /></font></div> | ||
| − | + | == Sun Raster (RS) files == | |
<div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> RS files is the native bitmap format for Sun Microsystems UNIX platforms that run the SunOS operating system. This format stores bitmap data (color, grayscale, black and white) of any pixel/voxel depth. RS files can be of any size; however, multiple images per file are not supported. The header is embedded in the file; it is 32 bytes in the length and contains typical header information such as the width and height of the image and the type of color map used.<br /></font></div> | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> RS files is the native bitmap format for Sun Microsystems UNIX platforms that run the SunOS operating system. This format stores bitmap data (color, grayscale, black and white) of any pixel/voxel depth. RS files can be of any size; however, multiple images per file are not supported. The header is embedded in the file; it is 32 bytes in the length and contains typical header information such as the width and height of the image and the type of color map used.<br /></font></div> | ||
| − | + | == Tag Image File Format (TIFF) files == | |
<div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> [http://en.wikipedia.org/wiki/TIFF TIFF is a bitmap file format that is generally used to provide a portable, image-storage mechanism that describes image data. Multiple images can be stored in one file. TIFF files can accommodate a maximum image size of 2]<span style="font-style: normal; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: super"><font color="#000000">32</font></span> -1 pixel/voxels. It is widely used and is a standard file format used in many desktop publishing, imaging, and paint programs. MIPAV supports most TIFF 6.0 formatted files used commonly in the image research community including 2D and 3D monochrome 8-bit, signed and unsigned 16-bit, and signed 32-bit images. Future support of 24-bit color and compressed images is planned. The TIFF Image File Header (IFH) is 8 bytes in length and contains 3 fields of information. If multiple images are in a file, an IFH is present for each image in the file.<br /></font></div> | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> [http://en.wikipedia.org/wiki/TIFF TIFF is a bitmap file format that is generally used to provide a portable, image-storage mechanism that describes image data. Multiple images can be stored in one file. TIFF files can accommodate a maximum image size of 2]<span style="font-style: normal; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: super"><font color="#000000">32</font></span> -1 pixel/voxels. It is widely used and is a standard file format used in many desktop publishing, imaging, and paint programs. MIPAV supports most TIFF 6.0 formatted files used commonly in the image research community including 2D and 3D monochrome 8-bit, signed and unsigned 16-bit, and signed 32-bit images. Future support of 24-bit color and compressed images is planned. The TIFF Image File Header (IFH) is 8 bytes in length and contains 3 fields of information. If multiple images are in a file, an IFH is present for each image in the file.<br /></font></div> | ||
| − | + | == TARGA == | |
<div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> [http://ozviz.wasp.uwa.edu.au/~pbourke/dataformats/tga/ TGA or TARGA format is a format for describing bitmap images, it is capable of representing bitmaps ranging from black and white, indexed color, and RGB color. See also "Creating TGA Image files" written by Paul Burke.]<br /></font></div> | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> [http://ozviz.wasp.uwa.edu.au/~pbourke/dataformats/tga/ TGA or TARGA format is a format for describing bitmap images, it is capable of representing bitmaps ranging from black and white, indexed color, and RGB color. See also "Creating TGA Image files" written by Paul Burke.]<br /></font></div> | ||
| − | + | == Truevision Graphics Adapter (TGA) files == | |
<div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> [http://en.wikipedia.org/wiki/TGA TGA is a bitmap file format that is often used in graphics, imaging, and paint applications that store up to 32-bit color images (8-, 16-, 24-, and 32-bit colors are supported). Created by Truevision, Incorporated, it is an uncompressed file format that does not support multiple images per file. The maximum image size however, is unlimited. TGA's header is 18 bytes in length and contains traditional header information including the depth of the color map entries.]<br /></font></div> | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> [http://en.wikipedia.org/wiki/TGA TGA is a bitmap file format that is often used in graphics, imaging, and paint applications that store up to 32-bit color images (8-, 16-, 24-, and 32-bit colors are supported). Created by Truevision, Incorporated, it is an uncompressed file format that does not support multiple images per file. The maximum image size however, is unlimited. TGA's header is 18 bytes in length and contains traditional header information including the depth of the color map entries.]<br /></font></div> | ||
| − | + | == VRML == | |
<div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> [http://www.web3d.org/x3d/specifications/vrml/vrml97/index.htm The Virtual Reality Modeling Language (VRML) is a file format for describing interactive 3D objects and worlds. It is based upon the OpenInventor file format which was originally developed by Silicon Graphics, Inc. For more information and format specifications, refer to <http://www.web3d.org/x3d/specifications/vrml/vrml97/index.htm>]<br /></font></div> | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> [http://www.web3d.org/x3d/specifications/vrml/vrml97/index.htm The Virtual Reality Modeling Language (VRML) is a file format for describing interactive 3D objects and worlds. It is based upon the OpenInventor file format which was originally developed by Silicon Graphics, Inc. For more information and format specifications, refer to <http://www.web3d.org/x3d/specifications/vrml/vrml97/index.htm>]<br /></font></div> | ||
| − | = | + | <div id="VtkXml"></div> |
| + | |||
| + | == VTK XML surface == | ||
<div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> [http://www.vtk.org/pdf/file-formats.pdf See VTK documentation available at <http://www.vtk.org/pdf/file-formats.pdf>.]<br /></font></div> | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> [http://www.vtk.org/pdf/file-formats.pdf See VTK documentation available at <http://www.vtk.org/pdf/file-formats.pdf>.]<br /></font></div> | ||
| − | + | == X BitMap (XBM) files == | |
<div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> XBM is a monochrome bitmap file format that is used primarily for the storage of icon and cursor bitmaps in X Windows. It was developed by X Consortium. Because this format was developed for small amounts of data, the bitmap images are composed of collections of ASCII data rather than binary data. (XBM bitmap data is often found in C source header files.) Multiple images can be stored on a file; there is no limit to the image size. XBM has no header file, nor is a formal header embedded in the program; rather header information consists of four lines that begin with <span style="font-style: normal; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline"><font color="#000000"><nowiki>#define.</nowiki></font></span> The four lines of code indicate the height and width of the image and the coordinate of the hotspot (if any).<br /></font></div> | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> XBM is a monochrome bitmap file format that is used primarily for the storage of icon and cursor bitmaps in X Windows. It was developed by X Consortium. Because this format was developed for small amounts of data, the bitmap images are composed of collections of ASCII data rather than binary data. (XBM bitmap data is often found in C source header files.) Multiple images can be stored on a file; there is no limit to the image size. XBM has no header file, nor is a formal header embedded in the program; rather header information consists of four lines that begin with <span style="font-style: normal; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline"><font color="#000000"><nowiki>#define.</nowiki></font></span> The four lines of code indicate the height and width of the image and the coordinate of the hotspot (if any).<br /></font></div> | ||
| − | + | == XML == | |
<div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> [http://en.wikipedia.org/wiki/Standard_Generalized_Markup_Language Extensible Markup Language (XML) is a very flexible markup language format derived from SGML. However, XML was originally designed primarily for large-scale electronic publishing, now it is playing an significant role in the exchange of a wide variety of data including image processing data. For more information about XML, refer to <][http://en.wikipedia.org/wiki/XML ]<span style="font-style: normal; font-weight: normal; text-transform: none; vertical-align: baseline"><u><font color="#000000">http://en.wikipedia.org/wiki/XML</font></u></span>>. <br /></font></div> | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> [http://en.wikipedia.org/wiki/Standard_Generalized_Markup_Language Extensible Markup Language (XML) is a very flexible markup language format derived from SGML. However, XML was originally designed primarily for large-scale electronic publishing, now it is playing an significant role in the exchange of a wide variety of data including image processing data. For more information about XML, refer to <][http://en.wikipedia.org/wiki/XML ]<span style="font-style: normal; font-weight: normal; text-transform: none; vertical-align: baseline"><u><font color="#000000">http://en.wikipedia.org/wiki/XML</font></u></span>>. <br /></font></div> | ||
| Line 386: | Line 422: | ||
MIPAV has it's own XML version - MIPAV XML, which is explained in Section [MIPAV_AppCSupportedFormats.html#1260832 "MIPAV XML" on page 571]. | MIPAV has it's own XML version - MIPAV XML, which is explained in Section [MIPAV_AppCSupportedFormats.html#1260832 "MIPAV XML" on page 571]. | ||
| − | + | == XML surface == | |
<div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> MIPAV defined XML surface format based on the MIPAV "surface.xsd" and "surfaceref.xsd" file. The XML surface file defines specific variables for reading and writing surface.xml files.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> MIPAV defined XML surface format based on the MIPAV "surface.xsd" and "surfaceref.xsd" file. The XML surface file defines specific variables for reading and writing surface.xml files.<br /></font></div><div style="font-style: normal; font-weight: normal; margin-bottom: 0pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> | ||
| Line 408: | Line 444: | ||
<br /></font></div> | <br /></font></div> | ||
| − | + | == X PixMap (XPM) files == | |
<div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> XPM is a bitmap file format developed by Groupe Bull. It is used primarily to store X Windows pixmap information. XPM can store black and white, grayscale or color image data of unlimited colors. Like XBM, there is no limit for the image size and there can be multiple images per file. In addition, like XBM, XPM is written in ASCII. It can contain a section, <span style="font-style: normal; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline"><font color="#000000"><Values></font></span>, that contains header information, such as the height and width of the pixmap and the number of characters per pixel or voxel.<br /></font></div> | <div style="font-style: normal; font-weight: normal; margin-bottom: 6pt; margin-left: 0pt; margin-right: 0pt; margin-top: 0pt; text-align: left; text-decoration: none; text-indent: 0pt; text-transform: none; vertical-align: baseline"><font color="#000000"> XPM is a bitmap file format developed by Groupe Bull. It is used primarily to store X Windows pixmap information. XPM can store black and white, grayscale or color image data of unlimited colors. Like XBM, there is no limit for the image size and there can be multiple images per file. In addition, like XBM, XPM is written in ASCII. It can contain a section, <span style="font-style: normal; font-weight: normal; text-decoration: none; text-transform: none; vertical-align: baseline"><font color="#000000"><Values></font></span>, that contains header information, such as the height and width of the pixmap and the number of characters per pixel or voxel.<br /></font></div> | ||
| + | == See also: == | ||
| + | |||
| + | *[[Supported Formats]] | ||
| + | **[[Supported Formats | Graphical and file formats supported by MIPAV]] | ||
| + | **[[MIPAV configuration files]] | ||
| + | **[[FAQ: Understanding Image Basics#FilesExtensions | Image files supported by MIPAV]] | ||
| + | *[[Understanding MIPAV-related files]] | ||
| − | [[ | + | [[Category: Help]] |
Latest revision as of 16:40, 13 January 2014
Contents
- 1 Adobe Photoshop (PSD) files
- 2 Analyze files
- 3 AFNI
- 4 Audio Video Interleave (AVI) files
- 5 BIORAD
- 6 Bruker data format
- 7 Cheschire (IMG or IMC)
- 8 Digital Imaging and Communications in Medicine (DICOM) files
- 9 FITS
- 10 FreeSurfer image and surface files
- 11 GE - Genesis 5X and LX
- 12 Graphics Interchange Format (GIF) files
- 13 ICS (Image Cytometry Standard)
- 14 Joint Photographic Experts Group (JPEG/JFIF) files
- 15 Laser Scanning Microscope (Zeiss)
- 16 Legacy VTK
- 17 LIFF
- 18 Macintosh PICT
- 19 Medical Image NetCDF (MINC) files
- 20 Medical Research Council (MRC)
- 21 MICRO-CAT
- 22 Microsoft Windows Bitmap (BMP) files
- 23 MIPAV (LUT) files
- 24 MIPAV (MTX) files
- 25 MIPAV (PLT) files
- 26 MIPAV text file format
- 27 MIPAV (SUR) files
- 28 MIPAV (VOI) files
- 29 MIPAV XML
- 30 MIPAV XML surface format
- 31 NIFTI
- 32 PC Paintbrush (PCX) files
- 33 PICT files
- 34 Phillips PAR/REC
- 35 POLY
- 36 Polygon File Format
- 37 Portable Network Graphic Format (PNG) files
- 38 QuickTime-Apple
- 39 Raw data files
- 40 Siemens - Magnetom Vision
- 41 STL (ASCII and Binary)
- 42 Sun Raster (RS) files
- 43 Tag Image File Format (TIFF) files
- 44 TARGA
- 45 Truevision Graphics Adapter (TGA) files
- 46 VRML
- 47 VTK XML surface
- 48 X BitMap (XBM) files
- 49 XML
- 50 XML surface
- 51 X PixMap (XPM) files
- 52 See also:
Adobe Photoshop (PSD) files
PSD is a bitmap file format created by Adobe. It can accommodate images with an unlimited amount of colors. PSD files are not multiple image files; the maximum image size is 30K x 30K pixels or voxels. PSD files are generally used to store images that were altered or manipulated by the Adobe Photoshop software. The header is embedded in the file and is 26 bytes in length. It contains information such as the height and width of the image and the color mode.
Analyze files
Analyze formatted files are generated by a UNIX-based, image-processing application developed at the Mayo Clinic. In support of some NIH intramural researchers who have used Analyze in the past and have generated a large number of legacy datasets, MIPAV reads and writes Analyze formatted images.
An image file in Analyze format contains uncompressed voxel data for the images in one of several possible voxel formats:
1 bit packed binary (slices begin on byte boundaries)
8 bit unsigned char (grayscale unless .lkup file is present)
16 bit signed short
32 bit signed integers or float
24 bit RGB, 8 bits per channel
The header file is a C structure that describes the dimensions and properties of the voxel data.
Analyze images are formed from the following files:
Header file - This file (extension .HDR) describes the image type, size, and other important image attributes. For example, brain.hdr is the header file that describes the image file.
File Containing the Actual Image Data - This file (extension .IMG) can be interpreted as a raw file since it does not have any header information within the file. For example, brain.img is the file that contains the image data.
A color lookup file is sometimes included in the file set - A LKUP extension indicates a lookup table file. Example: file.lkup.
All files from the file set have the same name and are distinguished by the extensions HDR for the header file, IMG for image file and LKUP for lookup table file. MIPAV stores files in Analyze format in the catalog you specified in the Save Image as dialog box.
Note: MIPAV could save images with *.img extension either in Analyze format or in NIFTI format. To save an image in Analyze format, call the File > Save Image as menu, and then type the file name with *.img extension. In the Choose Type of File to Write dialog box that appears, specify the file format (Analyze). Check the option Always save .img files in Analyze format, if you want MIPAV to save .img files only in that format.
For more information about Analyze file format, please refer to the ANALYZE 7.5. format document from the Mayo Clinic.
AFNI
The HEAD file for an AFNI dataset is in ASCII, so you can view it with a normal text editor (e.g., Notepad). The data within are organized into attributes, which are named arrays of floats, integers, or characters (strings). A sample float attribute is shown in Table 2. This array defines the voxel array dimensions. An example of a character array attribute is shown in Table 3.
|
type = integer-attribute name = DATASET_DIMENSIONS count = 5 256 256 124 0 0
|
|
type = string-attribute name = IDCODE_DATE count = 25 'Mon Apr 15 13:08:36 1996~
|
Note that the data for a string attribute starts with a single apostrophe (') character, and that the number of bytes is given by the count parameter. The ASCII NUL character is replaced by the tilde ~ so that the HEAD file can be edited manually, if need be. On input, tildes will be replaced with NULs.
The dataset reading code requires a minimal set of attributes to be able to decipher a dataset; for example, the DATASET_DIMENSIONS attribute above is required. There are also some attributes that are not mandatory for a dataset to be successfully constructed from a HEAD file; for example, the IDCODE_DATE attribute above is not required. Attributes that AFNI doesn't know how to deal with are ignored. This makes it possible for a program to tuck extra information into the HEAD file and not cause a trouble.
A BRIK file contains nothing but voxel values. If a dataset has 1 sub-brick, which is 100x100x100 voxels, and the values stored are shorts (2 bytes each), then the BRIK file will have exactly 2,000,000 bytes. All the formatting information is stored in the HEAD file.
Audio Video Interleave (AVI) files
BIORAD
Bruker data format
Bruker format stores a single scanning session in its own directory. The directory is named according to the subject name or number, as typed in by the scanner operator. The directory name usually specifies the subject name/number and which session this is for that subject. For more information, refer to <http://imaging.mrc-cbu.cam.ac.uk/imaging/FormatBruker>.
Cheschire (IMG or IMC)
Digital Imaging and Communications in Medicine (DICOM) files
The DICOM format is a standard that defines a standard method of communication between two devices, such as computers, servers, or imaging devices. DICOM not only prescribes how to communicate with other imaging equipment and databases but also specifies how images are stored. Each image has accompanying header information describing the image format (i.e., height, width, etc.) as well as information that indirectly relates to the image (i.e., patient information, image equipment setup parameters).
See also: Working with DICOM Images.
FITS
FITS stands for "Flexible Image Transport System" and is the standard astronomical data format endorsed by both NASA and the IAU. FITS is primarily designed to store scientific data sets consisting of multi-dimensional arrays (1-D spectra, 2-D images or 3-D data cubes) and 2-dimensional tables containing rows and columns of data.
For more information about FITS refer to http://heasarc.gsfc.nasa.gov/docs/heasarc/fits.html.
FreeSurfer image and surface files
FreeSurfer uses two main kinds of data. The first one is a volume of voxels, as for example, from an MRI scanner. It provides a source of raw input. The other one is surface data which consist of lists of vertices as well as their positions in space and faces and also vertices associated with them.
For more information, refer to Surface file formats web.
GE - Genesis 5X and LX
The format of the headers is General Electric Genesis described in the [Medical Image Format FAQ]. The image header contains the table position and field of view. This is important information, because the spacing between the slices and the pixel size changes several times throughout the data set.
The slices are named as follows: c_vmxxxx.fre where xxxx is the location in mm's of the slice. There are 522 slices in the fresh CT data set, which requires ~ 274 megabytes of disk storage.
See also:
Graphics Interchange Format (GIF) files
The Graphics Interchange Format (GIF) is a bitmap file format that was created by CompuServe, Inc. GIF is primarily an exchange and storage file format; GIFs can be used to store one or several bitmap images in one file. GIFs support pixel or voxel depths of 1 to 8 bits. The maximum image size is 64K x 64K pixel or voxels. There are two major revisions of the GIF format specification: GIF87a and GIF89a. Both formats begin with a 6-byte header that identifies the file format as GIF.
ICS (Image Cytometry Standard)
The Image Cytometry Standard (ICS) is a digital multidimensional image file format used in life sciences microscopy. It stores the image data and the microscopic parameters describing the optics during the acquisition. The newest ICS2 file format uses a single ICS file with both the header and the data together.
The ICS format is capable to store: multidimensional and multichannel data images in 8, 16 or 32 bit integer, 32 or 64 bit floating point, floating point complex data, and all microscopic parameters directly relevant to the image formation free-form comments.
For more information, refer to http://en.wikipedia.org/wiki/Image_Cytometry_Standard.
Joint Photographic Experts Group (JPEG/JFIF) files
Laser Scanning Microscope (Zeiss)
Laser Scanning Microscope (LSM) is a line of laser point scanning microscopes produced by Carl Zeiss LLC. The new LSM 700/710/780 series were released after 2008, thus making the 5th generation, including the LSM 510, LSM 5 Pascal, and LSM 5 Live obsolete.
LSM models use file format with the filename extension ".lsm". Particular format specification depends on the microscope model, but all are essentially extensions of the TIFF multiple image stack file format, containing instrument specific hardware settings metadata and thumbnail images.
For format details, refer to http://en.wikipedia.org/wiki/LSM_(Zeiss).
Legacy VTK
The legacy VTK file format is a surface format that consist of five basic parts:
- The file version and identifier, it contains the single line: # vtk DataFile Version x.x. This line must be exactly as shown with the exception of the version number x.x, which will vary with different releases of VTK.
- The header, it consists of a character string terminated by end-of-line character \n. The header is 256 characters maximum. The header can be used to describe the data and include any other pertinent information.
- The file format, which describes the type of file, either ASCII or binary. On this line the single word ASCII or BINARY must appear. 4 The dataset structure that can have the geometry section and actual data. The geometry section describes the geometry and topology of the dataset. This part begins with a line containing the keyword DATASET followed by a keyword describing the type of dataset. Then, depending upon the type of dataset, other keyword/data combinations define the actual data.
- The dataset attributes. It begins with the keywords POINT_DATA or CELL_DATA, followed by an integer number specifying the number of points or cells, respectively. Other keyword/data combinations then define the actual dataset attribute values (i.e., scalars, vectors, tensors, normals, texture coordinates, or field data).
Note: the current version number is 3.0. Version 1.0 and 2.0 files are compatible with version 3.0 files.
For more information about VTK file formats, refer to VTK file formats.
LIFF
Layered Image File Format is a file format used in the Openlab suite for microscope image processing. It is a proprietary format, but has an open, extensible form analogous to TIFF. It was specifically designed to contain a large number of high resolution images, and also all of the meta data generated by analysis of such images.
For more information, refer to OpenLab web site.
Macintosh PICT
The Macintosh PICT (Macintosh Picture) format is format for the Macintosh. It also known as PICT, Macintosh Picture, or QuickDraw Picture Format. All Macintosh PICT files start with a 512-byte header, which contains information that the Macintosh uses to keep track of the file. This is followed by three fields describing the image size (picSize), the image frame (picFrame), and a version number. For more information, refer to the Macintosh PICT File Format Summary page at <http://www.fileformat.info/format/macpict/egff.htm>.
Medical Image NetCDF (MINC) files
MINC is a medical-imaging file format that is based on the Network Common Data Form (NetCDF) file format. NetCDF is a platform-independent software interface that provides a means for storing named, multidimensional variables. Each multidimensional variable is defined by a name, dimensions, and attributes. For example, cardio (name) [256, 256] (dimensions), and "long_name" where this attribute is a string that describes the content of the image. MINC provides a standard for dimension, variable, and attribute names suitable for medical imaging. MINC also provides convenience functions to complement the NetCDF interface and convenience functions for using MINC files.
For more information, refer to the MINC web page.
Medical Research Council (MRC)
The MRC file format is a file format for electron density. It is an industry standard for Cryo-electron microscopy where the result of the technique is a three-dimensional grid of voxels each with a value corresponding to density of electrons. The MRC format is supported by almost every molecular graphics suite that supports volumetric data.
For more information, refer to this doc.
MICRO-CAT
Siemens microCAT image format is designed for its microCAT+SPECT high resolution, high sensitivity, in vivo preclinical imaging system, which allows detect and quantify anatomical and functional changes in animals associated with disease.
For more information refer to: Siemens press release.
Microsoft Windows Bitmap (BMP) files
BMP is a native bitmap format for the Microsoft Windows and OS/2 platforms. Developed by the Microsoft Corporation, it can be read on Intel machines running Microsoft Windows, Windows XP, Windows NT, Windows 95, OS/2, and MS-DOS. It is an uncompressed file format with a maximum image size of 32K x 32K or 2G x 2G pixels or voxels (depending on the version of BMP).br />
Depending on the version of BMP, for OS/2 the maximum image size is larger, 64K x 64K or 4G x 4G pixels or voxels. There are several versions of BMP for Microsoft Windows and OS/2. MIPAV supports Microsoft Window BMP versions 2.x and above and all OS/2 versions of the BMP file format. BMP version 2.x is designed for use with the Microsoft Windows 2.x platform. It has a 14-byte header (as does the OS/2 1.x bitmap header). It can accommodate images with 1-, 4-, 8-, or 24-bit colors. Version 3.x is designed for use with the Microsoft Windows 3.x and Windows NT platforms. Like version 2.x, it contains a 14-byte header. It also contains an additional bitmap header that is 40 bytes in size. The Microsoft Windows 3.x platform version accommodates images with 1-, 4-, 8- or 24-bit colors. Windows NT and Windows XP accommodate 16- and 32-bit images. Version 4.x was designed for use with Microsoft Windows 95. It contains the 14-byte header, and an additional 108-byte bitmap header. It can accommodate images of 1, 4, 8, 16, and 32 bits.
MIPAV (LUT) files
LUT is a vector file format used to store MIPAV Lookup Table ( LUT) data.
MIPAV provides a logical color map, which allows you to remap the original intensities to other intensities. Although technically the term look-up table (LUT) can be used for the physical and logical color maps, in this guide look-up table refers to the logical color map only. You can apply predefined, pseudo color or inverse LUTs, or you can manually manipulate the transfer function used to map the image data to the LUT. The LUT then translates the remapped values so that they can be interpreted by the physical color map and displayed on your monitor.
For a sample LUT file refer to LUT file.
MIPAV (MTX) files
The matrix filedescribes the image's orientation, translations, offset scales, rotations, and shears. When any of this information changes in the image, the identity matrix changes.
For a sample .MTX file refer to Matrix file.
MIPAV (PLT) files
The MIPAV plot file (.PLT) is a vector file that is used to store graphics data. For a sample PLT file refer to Plot file.
MIPAV text file format
MIPAV defined ASCII text surface format. The text surface file records the triangle mesh vertices, normals and connectivity.
Vertices x, y, z // vertex position
Normal
x, y, z // normal coordinate
Connectivity
x, y, z // index connection
See also: MIPAV surface file in text format.
MIPAV (SUR) files
MIPAV supports its own surface format (*.sur). A MIPAV surface file contains the information about the surface area and geometry, plus the additional information that describes the surface color and more. This is defined by an XML scheme, which can be found on the MIPAV web site.
Surface format (Triangle mesh or ClodMesh - Continuous level of details mesh)
Inverse Dicom matrix flag
3D mesh direction x, y, z
Start location x, y, z
Bounding box x, y, z
Inverse Dicom matrix
Vertex quantity
Vertex position x, y, z
Vertex normal x, y, z
Index quantity
Index connectivity x, y, z
Surface material property
Clodmesh collapse record
See also MIPAV Surface files description.
MIPAV (VOI) files
VOI is a vector file format that is used to store volume of interest contouring information. For a sample file refer to [MIPAV_AppCSupportedFormats.html#1249118 "VOI file" on page 603].
MIPAV XML
Refer to MIPAV XML.
MIPAV image schema definition [1]
MIPAV XML surface format
See also MIPAV Image Schema.
MIPAV Image Schema can also be found here.
NIFTI
NifTy-1 is a new Analyze-style data format, proposed by the NIfTI DFWG. NIfTI-1 was adapted from ANALYZE 7.5 file format. See also NIFTI.
It uses the "empty space" in the ANALYZE 7.5 header to add several new features, which are listed below:For more information, refer to the NIfTI web site at <http://nifti.nimh.nih.gov/>.
PC Paintbrush (PCX) files
PC Paintbrush is a bitmap file format that is used primarily in Microsoft Windows and other Windows-based products. PCX is mainly used as an exchange and storage format. Created by ZSoft and packaged with Microsoft Windows, PCX can accommodate 1-, 4-, 8-, and 24-bit color images. It is uncompressed; the maximum image size is 64K x 64K pixel/voxels. Header information is embedded in the file; the first 128 pixel/voxels in the file contain information such as the PCX ID number, the bits per pixel/voxel, and the palette type.
PICT files
See also Supported file formats - PICT.
Phillips PAR/REC
See also <http://www.dclunie.com/medical-image-faq/html/part8.html>
and MIPAV User Guide, Volume 2, Algorithms "DTI Create List File".
POLY
Below, is the description of POLY format originally written in 1993 by Pat Flynn and revised 1/96 PJF. For more information refer to:
Each vertex appears on its own line.
The format is `pnt %d: %f %f %f'.
Points are numbered starting at zero. There is no sentinel indicating the end of the points and the beginning of the polygons; your program logic can easily detect it.
Each polygon appears on its own line.
The format is `poly %d: [%d] %d %d %d .... ' The number on the left of the colon is the index of the polygon (numbered from zero). The number inside the [square brackets] is the number of vertices. The remaining numbers are the indices of the vertices. The vertices proceed clockwise around the polygon as you look at it from `outside' the object.
The surface list allows to logically group polygons.
The format for a surface entry is:
Polygon File Format
|
format ascii 1.0 { ascii/binary, format version number } comment made by Greg Turk { comments keyword specified, like all lines } element vertex 8 { define "vertex" element, 8 of them in file } property float x { vertex contains float "x" coordinate } property float y { y coordinate is also a vertex property } property float z { z coordinate, too } element face 6 { there are 6 "face" elements in the file } property list uchar int vertex_index { "vertex_indices" is a list of ints } end_header { delimits the end of the header } 0 0 0 { start of vertex list } 0 0 1 0 1 1 0 1 0 1 0 0 1 0 1 1 1 1 1 1 0 3 1 2 3 { start of face list } 3 6 5 4 3 4 5 1 3 5 6 2 3 6 7 3 3 7 4 0
|
Portable Network Graphic Format (PNG) files
QuickTime-Apple
See also: QuickTime-Apple.
Raw data files
Siemens - Magnetom Vision
Refer to http://www.dclunie.com/medical-image-faq/html/part4.html#MagnetomVision and Siemens - Magnetom Vision.
STL (ASCII and Binary)
Sun Raster (RS) files
Tag Image File Format (TIFF) files
TARGA
Truevision Graphics Adapter (TGA) files
VRML
VTK XML surface
X BitMap (XBM) files
XML
MIPAV has it's own XML version - MIPAV XML, which is explained in Section [MIPAV_AppCSupportedFormats.html#1260832 "MIPAV XML" on page 571].