DTI Pipeline
This article is a stub. It needs improvement.
Introduction
The DTI pipeline of MIPAV prepares diffusion weighted images (DWIs) and computes voxel-wise diffusion tensors (DT) for the further analysis of diffusion tensor imaging (DTI) data, see MIPAV DTI Color Display. The pipeline computes maps of diffusion eigenvalues and eigenvectors. It also determines an anatomical correspondence between DTI and structural MRI images of the same sample.
MIPAV DWI pipeline overview
This section is correct.
Brain MRI introduction
Brain MRI is usually performed with a 1.5- or 3- T MRI machine, with a gradient strength in the range of 20-60 mT/m and a slew rate of 120 T/m/s. Parameters for a single-shot spin-echo echo-planar imaging (EPI) sequence include: a repetition time (TR) of 6000 ms, an echo time (TE) of 100 ms, a field of view (FOV) of 24 cm. These parameters are typically used to obtain 3- to 5-mm axial or coronal sections with a 5-mm intersection gap. The acquisition matrix is usually 96 × 96 with a reconstruction matrix set to 128 × 128. The DWIs are obtained by using 4 linearly increasing b values in 6-7 non-collinear directions (bmax ~ 703-1000 s/mm2). In addition, a T2-weighted (T2W) reference image is obtained without diffusion weighting. Read more: [1], [2].
Determining an anatomical correspondence between DTI and structural MRI images of the same sample
MIPAV uses an image-based registration scheme mainly because i) it doesn't require a field map, which is usually not available for DWIs, and ii) it allows one to correct for mis-registration produced by a patient motion. For more information, refer to [3], [4].
To measure how well the images are spatially aligned MIPAV uses one of the cost functions, e.g., Correlation Ratio, Least Squares, Normalized Cross Correlation, or Normalized Mutual Information.
A T2 image (uploaded by a user) is chosen as a reference for all other images in the dataset. We use the T2 as a target image because it is usually less distorted and has a higher signal-to-noise ratio (SNR) than DWIs. Then, using a spatial transformation model, MIPAV aligns all other images to the target image by optimizing the cost function.
Note: In MIPAV, we use the term cost function to refer to the negative cost function.
MIPAV DTI pipeline outline
- A user uploads a DWI image and T2 image to the pipeline using the Import Data panel. A DWI image can be acquired from many different MRI scanners (including Philips, Siemens, GE, etc.) and in various formats. MIPAV reads the gradient information from the image header, or from the B-matrix file uploaded by the user. The gradient (or B-matrix) information is then displayed in the Gradient table. For the list of image types and how MIPAV reads the header information, refer to Image Types section.
- In the Pre-processing step, the B0 slice in DWI is detected, and then rigidly registered to the T2 image. The DW image is then registered to rigidly aligned B0 using the Optimized Automatic Registration 3.5 D algorithm. These steps are necessary to perform a motion correction and eddy current distortion correction.
- In the EPI Distortion Correction step, MIPAV calculates deformation vector fields for rigidly aligned B0 and T2, which came from the Pre-processing step. MIPAV then uses both: the transformation matrices obtained in the Pre-processing step, and deformation vector field values to create a corrected DWI image.
- MIPAV then creates a tensor using pre-processing DWI and the gradient/B-value information and a mask image uploaded by the user.
- MIPAV uses the tensor information to create a whole bunch of images, including ADC, color map, Eigen value, Eigen vector, FA, RA, and Volume Ratio.
- MIPAV creates a 3D visualization of fiber bundle tracts in the brain's white matter using the information from the previous step. The user can save fiber tracts information as .vtk and .dat files. See also: Image formats descriptions.
Image types
| Image file type | Auto Population of Bvals | Auto Population of Gradients | Auto Population of Bmatrix | Philips Gradient Creator Utility |
|---|---|---|---|---|
| Philips PAR/REC V3 &V4 | Yes | Yes | No | Yes |
| Philips PAR/REC 4.1 &4.2 | Yes | Yes | No | Yes |
| Philips DCM V3 &V4 | Yes | Yes | No | Yes |
| Philips DCM 4.1 &4.2 | Yes | Yes | No | Yes |
| Siemens Mosaic DCM | No | No | Yes | No |
| Nifti w/ Philips Par File | Yes | Yes | No | Yes |
| GE DCM | In progress | In progress | In progress | In progress |
| Text File Type | Auto Population of Bvals | Auto Population of Gradients | Auto Population of Bmatrix | Philips Gradient Creator Utility |
| fslBvalGrad.txt | Yes | Yes | No | No |
| dtiStudioBvalGrad.txt | Yes | Yes | No | No |
| mipavStandardBvalGrad.txt | Yes | Yes | No | No |
| dcm2nii.bvec | Yes | Yes | No | No |
| fslBmatrix.txt | No | No | Yes | No |
| mipavStandardBmatrix.txt | No | No | Yes | No |
MIPAV DTI Pipeline dialog box
Import Data tab
This tab is for uploading DW and T2 images, calculating B-values and creating B-matrix (Gradient) table.
Upload DWI Image box
DTI Pipeline reads raw data, all [[Supported Formats | MIPAV supported formats] and DICOM files.
DWI Image Browse – check this option if you would like to upload your image of interest from your computer.
Use Active DWI Image – check this option to use an active image, which is already opened in MIPAV.
Note: in MIPAV, an active image is the one that has a red frame.
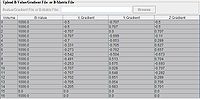
Upload B-Value/Gradient File or B-Matrix file
This options allows a user to manually upload B-Value/Gradient File or B-Matrix file.
Introducing B-value: - a diffusion gradient can be represented as a 3D vector q. The direction of vector q is in the direction of diffusion and its length is proportional to the gradient strength. The gradient strength, or more often used diffusion weighting parameter, is s expressed in terms of the b value parameter, which is proportional to the product of the square of the gradient strength q and the diffusion time interval (b ~ q2 • Δt). Read more: MRI TIP database.
Gradient creator
The Gradient Creator computes the correct gradient table which is necessary to calculate diffusion tensors. The table lists gradient vectors that describe the diffusion weighting directions, which are later used for analysis and computing diffusion tensor. The Gradient Creator works with data acquired by Philips MRI scanners.
It uses the source code from DTI_gradient_table_creator created by Jonathan Farrell, Ph.D.
The code that computes the gradient table based on minimal user input. The list of input parameters is show below, see Gradient Creator input parameters. You can also refer to Jonathan Farrell's web site for additional information.
The specifications of the gradient table created for the image taken on Philips MRI scanner(s) are determined by:
- The 3 coordinate frames defined on the MR scanner: world, anatomical, and image coordinate system. For more information, refer to Slicer WIKI.
- The rules on how imaging options (including slice orientation, slice angulation, phase encoding direction, etc.) impact the gradient directions.
- The scanner software release.
- Any motion correction, if performed.
Slice angulation
In diffusion tensor imaging (see [5]), tensors are constructed by collecting a series of direction-sensitive diffusion images. MRI scanners save these directions along with the images and later they are used to reconstruct the diffusion properties of the images.
Depending to the scanner, the vectors are recorded with reference either to the scanner bore, or to the imaging grid. This should not a problem if the images are acquired precisely orthogonal to the scanner bore, because in that case the image and the scanner share the same frame of reference.
Problems can arise in oblique acquisitions when the image plane is not aligned with the scanner bore. In this situation, it is important that the gradient vectors used in the imaging software are in the same frame of reference as the image. This requires conversion or re-ordering, otherwise we will get angulation errors.
Note: these angulation errors have little influence on the DTI parameters, which are invariant to tensor rotation - ADC, MD and FA. However, they do affect calculation of the eigenvectors of the tensor.
The Gradient or B-Matrix table
In the Gradient or B-Matrix table the B column contains b values, while X Gradient, Y Gradient and Z gradient columns contain the diffusion gradients applied along the x, y, and z axis - Gx, Gy, and Gz correspondingly.
Gradient creator input parameters
| Gradient Creator Parameters | Options | Philips PAR/REC Version |
|---|---|---|
| Fatshift | R: Right L: Left A: Anterior P: Posterior H: Head F: Feet |
V3 & V4 V4.1 & V4.2 |
| Jones30 or Kirby | Specify which MRI scanner was used to acquire DW images. This option works for KKI scanners only. | V3 & V4 V4.1 & V4.2 |
| Gradient Resolution | Low: used for 8 DWI volumes Medium: used for 17 DWI volumes High: used for 34 DWI volumes |
V3 & V4 V4.1 & V4.2 |
| Gradient Overplus | Yes No |
V3 & V4 V4.1 & V4.2 |
| Philips Release | Rel_1.5 Rel_1.7 Rel_2.0 Rel_2.1 Rel_2.5 Rel_11.x |
V3 & V4 V4.1 & V4.2 |
| Patient Position | Head First Feet First |
V3 & V4 |
| Patient Orientation | SP: Supine PR: Prone RD: Right Decubitus LD: Left Decubitus |
V3 & V4 |
| Fold Over | AP: Anterior-Posterior RL: Right-Left FH: Head-Feet/ Superior-Inferior |
V3 & V4 |
| OS (Operating System) | Windows VMS |
V3 & V4 V4.1 & V4.2 (for KKI scanners only) |
| Inverted | No Yes |
V3 & V4 V4.1 & V4.2 (for KKI scanners only) |
Specials notes
For more information regarding the importing DW images, refer to Jonathan Farrell's web site - http://godzilla.kennedykrieger.org/~jfarrell/software_web.htm.
- If your DTI data was acquired on a Philips MR Scanner with new software (release 1.5, 1.7, 2.0 or 2.1), the PAR file version may be V4.1. You can verify this by looking at the top right hand corner of the file for the following tag: Research image export tool V4.1. The other known versions are V3 or V4.
- The V4.1 PAR files list the diffusion weighting directions, however, these directions are provided not in the image space (see Slice angulation). Specifically, in the case the Gradient Overplus option is set to YES, the directions need to be rotated and corrected for slice angulation. This is automatically done by the Gradient Table creator.
- The Gradient Table Creator does not read gradient directions from the PAR file because this information is not available for all PAR files (V3 or V4). It uses the directions documented in the Philips source code instead.
Re-Ordering data (from Camino)
Probably the most common task is to re-order data from image to voxel order. In scanner order or image order, multi-component images are stored as consecutive volumes. This is convenient for visualization, since you can easily render a particular 3D volume. It is inconvenient for parallel processing, as you must read the entire 4D image in order to get the components for a particular processing. Data in voxel order stores all components for a particular voxel together. Thus you can read the image one voxel at a time, or skip ahead to particular voxel, without reading the entire image into memory. Camino does most of its I/O in voxel order. You can get into and out of scanner order with the scanner2voxel and voxel2scanner commands. Since these commands expect to deal with mostly raw data, they read and write floats by default (see below). You can change this behaviour with -inputdatatype and -outputdatatype options. For example:
scanner2voxel -voxels 983040 -components 60 -inputfile ScannerOrder.img -inputdatatype short > VoxelOrder.Bfloat where -components specifies the number of volumes in the 4D input and -voxels specifies the number of voxels (ie, X×Y×Z, where X, Y, Z are the dimensions of the image).