|
MIPAV>Olgavovk |
| Line 1: |
Line 1: |
| Diffusion tensor imaging (DTI) analyzes the tissues that have an internal fibrous structure, which is analogous to the anisotropy of some crystals (e.g.neural axons of white matter or muscle fibers in the heart). In those tissues, the diffusion of water displays anisotropy in certain directions. That means that molecules of water diffuse more rapidly in the direction aligned with the fibrous structure, and more slowly in the direction perpendicular to it. Thus, the measured rate of diffusion will differ depending on the fiber direction and an observer's point of view.
| | ''Main article: [[DTI Pipeline]].'' |
|
| |
|
| In diffusion-weighted imaging (DWI), at least 3 gradients are applied in 3 different directions, which is sufficient to estimate the trace of the diffusion tensor. From the diffusion tensor, diffusion anisotropy measures such as the fractional anisotropy (FA) eigenvalues and eigenvectors can be computed and displayed. The principal direction of the diffusion tensor can be also used [[DTI pipeline interface#VisualizationTab |to visualize the white matter connectivity of the brain]]. DTI has been proven to be very useful to diagnose vascular strokes in the brain and in other clinical applications.
| | This is detailed tutorial(s) on how to use MIPAV [[DTI Pipeline]]. The page is currently under construction, but soon it will be populated with following tutorials: |
|
| |
|
| '''Note:''' this diffusion model is a rather simplified model of the diffusion process. It assumes that diffusion within each image voxel is linear and homogeneous.
| | == Importing Philips PAR/REC files == |
|
| |
|
| == Introduction ==
| | [[File:TutorialPAR_REC_ImportDataTab.jpg|400px|thumb|right|The Import Data panel]] |
| The DTI pipeline of MIPAV prepares [http://www.mr-tip.com/serv1.php?type=db1&dbs=Diffusion%20Tensor%20Imaging diffusion weighted images (DWIs)] and computes voxel-wise diffusion tensors ([http://en.wikipedia.org/wiki/Diffusion_MRI#Diffusion_tensor_imaging DT]) for the further analysis of diffusion tensor imaging (DTI) data, see [[DTI Color Display| MIPAV DTI Color Display]]. The pipeline computes maps of diffusion [http://en.wikipedia.org/wiki/Diffusion_MRI#Measures_of_anisotropy_and_diffusivity eigenvalues] and [http://en.wikipedia.org/wiki/Diffusion_MRI#Measures_of_anisotropy_and_diffusivity eigenvectors]. It also determines an anatomical correspondence between DTI and structural MRI images (T2) of the same sample.
| | <ol> |
| | <li>Open a PAR/REC file of interest in MIPAV. Refer to [[Opening and loading image files]] if you are working with MIPAV for [[Getting Started Quickly with MIPAV | the first time]].</li> |
|
| |
|
| == MIPAV DWI pipeline overview ==
| | <li>In the Main menu, go to System Analysis > DTI Pipeline.</li> |
|
| |
|
| [[File:DtiPipelineWikiSmall.jpg |400px|thumb|right|MIPAV DTI Pipeline schematic]]
| | <li>In the Import Data panel that appears, check the Use Active DWI box.</li> |
|
| |
|
| === Brain MRI introduction ===
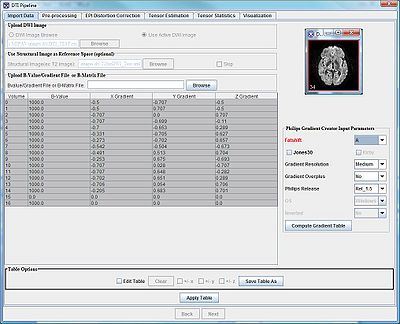
| | <li>Then, upload the file that contains gradient information. MIPAV reads the gradient information from the image header, or it can read it from the B-matrix file uploaded by the user. The gradient (or B-matrix) information is then populated in the Gradient table. </li> |
| Brain MRI is usually performed with a 1.5- or 3- T MRI machine, with a gradient strength in the range of 20-60 mT/m and a slew rate of 120 T/m/s. Parameters for a single-shot spin-echo echo-planar imaging (EPI) sequence include: a repetition time (TR) of 6000 ms, an echo time (TE) of 100 ms, a field of view (FOV) of 24 cm. These parameters are typically used to obtain 3- to 5-mm axial or coronal sections with a 5-mm intersection gap. The acquisition matrix is usually 96 × 96 with a reconstruction matrix set to 128 × 128. The DWIs are obtained by using 4 linearly increasing b values in 6-7 non-collinear directions (bmax ~ 703-1000 s/mm2). In addition, a T2-weighted (T2W) reference image is obtained without diffusion weighting. Read more: [http://www.ajnr.org/content/26/6/1455.full], [http://radiology.rsna.org/content/217/2/331.full].
| |
|
| |
|
| === Determining an anatomical correspondence ===
| | '''Note:''' for the full list of image types and how MIPAV reads the header information, refer to [[DTI Pipeline#ImageTypes | Image Types]] section. |
| between DTI and structural MRI images of the same sample. In MIPAV we use [http://ieeexplore.ieee.org/xpls/abs_all.jsp?arnumber=941749&tag=1 an image-based registration scheme] mainly because i) it doesn't require [http://www.mr-tip.com/serv1.php?type=db1&dbs=High%20Field%20MRI a field map], which is usually not available for DWIs, and ii) it allows one to correct for artifacts produced by a patient motion. For more information, refer to [http://ee.sharif.edu/~miap/Files/Medical%20Image%20Registration.pdf], [http://www.sciencedirect.com/science/article/pii/S0165027007002270].
| |
|
| |
|
| At least one of the following images is required as a reference in DTI pipeline:
| | <li> In the [[DTI pipeline interface#GradientCreator | Philips Gradient Creator Parameters box]], adjust the gradient parameters. Note that MIPAV can read the image information and will display an error message(s) if a parameter(s) is set to a wrong value. Refer to the [[DTI pipeline interface#GradientCreatorInput | Philips Gradient Creator Input parameters section]] for more information. And sample error messages and ways to put things right can be found in the [[DTI pipeline interface#ErrorMessages | Error Messages]] section.</li> |
|
| |
|
| *'''[http://www.mr-tip.com/serv1.php?type=db1&dbs=T2%20Weighted%20Image T2 image]''' - MIPAV uses the T2 as a reference image because it is usually less distorted and has a higher signal-to-noise ratio (SNR) than DWIs.
| | <li> Press Compute Gradient Table to apply new parameters.</li> |
| | <li> Press Next to proceed to the Pre-processing panel.</li> |
|
| |
|
| *'''B0''' - which is a DWI volume with no gradient applied. It can be used instead of T2, if T2 is not available.
| | '''Note:''' There is also the [[DTI pipeline interface#TableOptions | Table options box]] at the bottom of the panel. It allows the user to directly edit the values in the the Gradient table and to save the updated table. To edit the table, press the Edit button. To save the table, press Save Table As. To apply the changes, press Apply Table. The table could be save in various [[Supported Formats | formats]], including Plain text (.txt), FSL, dtistudio and MIPAV standard format. |
|
| |
|
| MIPAV aligns all other images to the reference image (either T2 or B0 volume) by optimizing [[Cost functions used in MIPAV algorithms | the cost function]], which represents the measure of how well the images are spatially aligned.
| |
|
| |
| '''Note:''' In MIPAV, we use the term "cost function" to refer to [[Cost functions used in MIPAV algorithms |the negative cost function]].
| |
|
| |
| === MIPAV DTI pipeline outline ===
| |
| <ol>
| |
| <li>The user [[DTI pipeline interface#ImportData | uploads a DWI image]] and [http://en.wikipedia.org/wiki/Spin-spin_relaxation_time T2 image] to the pipeline using the Import Data panel. A DWI image can be acquired from many different MRI scanners (including [http://www.healthcare.philips.com/us_en/products/mri/ Philips], [http://www.medical.siemens.com/webapp/wcs/stores/servlet/CategoryDisplay~q_catalogId~e_-1~a_categoryId~e_12754~a_catTree~e_100010,1007660,12754~a_langId~e_-1~a_storeId~e_10001.htm Siemens], [http://www.gehealthcare.com/euen/mri/index.html GE], etc.) and in [[Supported Formats | various formats]]. MIPAV reads the gradient information from the image header, or from the B-matrix file uploaded by the user. The gradient (or B-matrix) information is then displayed in the Gradient table. For the list of image types and how MIPAV reads the header information, refer to [[#ImageTypes | Image Types]] section.</li>
| |
|
| |
| <li>In the Pre-processing step, the B0 volume in DWI is detected (or entered by user), and then rigidly aligned to the T2 image. The DW image is then registered to rigidly aligned B0 using the[[Optimized automatic registration 3D | Optimized Automatic Registration 3.5 D]] algorithm. These steps are necessary to perform a motion correction and eddy current distortion correction.</li>
| |
|
| |
| <li>In the EPI Distortion Correction step, MIPAV calculates deformation vector fields for rigidly aligned B0 and T2, which came from the Pre-processing step. MIPAV then uses both: the transformation matrices obtained in the Pre-processing step, and deformation vector field values to create a corrected DWI image.</li>
| |
|
| |
| <li>MIPAV then calculates a diffusion tensor using pre-processing DWI and the gradient/B-value information and a mask image uploaded by the user.</li>
| |
|
| |
| <li>MIPAV uses the tensor information to create a tensor statistics, including ADC, color map, eigenvalue, eigenvector, FA, RA, and volume ratio images.</li>
| |
|
| |
| <li>MIPAV creates a 3D visualization of fiber bundle tracts in the brain's white matter using the information from the previous step. The user can save fiber tracts information as <span style="font-family:courier">[[Image formats descriptions#VtkXml |.vtk]]</span> and <span style="font-family:courier">[http://surfer.nmr.mgh.harvard.edu/fswiki/FsTutorial/Diffusion .dat]</span> files. See also: [[Image formats descriptions]].
| |
| </ol> | | </ol> |
|
| |
|
| <div id="ImageTypes"></div>
| | == Importing Siemens Mosaic DCM images == |
|
| |
|
| == Image types ==
| | TBD. |
|
| |
|
| {|style="color:black; background-color:#F8F8F8;" border="1" cellpadding="10" class="wikitable"
| | == Imploring Nifti with Philips Par File == |
| ! align="left"| Image file type
| |
| ! align="left"| Auto Population of Bvals
| |
| ! align="left"| Auto Population of Gradients
| |
| ! align="left"| Auto Population of Bmatrix
| |
| ! align="left"| Philips Gradient Creator Utility
| |
| |-
| |
| |Philips PAR/REC V3 &V4
| |
| |Yes
| |
| |Yes
| |
| |No
| |
| |Yes
| |
| |-
| |
| |Philips PAR/REC 4.1 &4.2
| |
| |Yes
| |
| |Yes
| |
| |No
| |
| |Yes
| |
| |-
| |
| |Philips DCM V3 &V4
| |
| |Yes
| |
| |Yes
| |
| |No
| |
| |Yes
| |
| |-
| |
| |Philips DCM 4.1 &4.2
| |
| |Yes
| |
| |Yes
| |
| |No
| |
| |Yes
| |
| |-
| |
| |Siemens Mosaic DCM, see also [[#ImportDataOutput | output files]]
| |
| |No
| |
| |No
| |
| |Yes
| |
| |No
| |
| |-
| |
| |Nifti w/ Philips Par File
| |
| |Yes
| |
| |Yes
| |
| |No
| |
| |Yes
| |
| |-
| |
| |GE DCM
| |
| |Yes
| |
| |Yes
| |
| |No
| |
| |No
| |
| |-
| |
| !align="left"|Text File Type
| |
| !align="left"| Auto Population of Bvals
| |
| !align="left"| Auto Population of Gradients
| |
| !align="left"| Auto Population of Bmatrix
| |
| !align="left"| Philips Gradient Creator Utility
| |
| |-
| |
| |fslBvalGrad.txt
| |
| |Yes
| |
| |Yes
| |
| |No
| |
| |No
| |
| |-
| |
| |dtiStudioBvalGrad.txt
| |
| |Yes
| |
| |Yes
| |
| |No
| |
| |No
| |
| |-
| |
| |mipavStandardBvalGrad.txt
| |
| |Yes
| |
| |Yes
| |
| |No
| |
| |No
| |
| |-
| |
| |dcm2nii.bvec
| |
| |Yes
| |
| |Yes
| |
| |No
| |
| |No
| |
| |-
| |
| |fslBmatrix.txt
| |
| |No
| |
| |No
| |
| |Yes
| |
| |No
| |
| |-
| |
| |mipavStandardBmatrix.txt
| |
| |No
| |
| |No
| |
| |Yes
| |
| |No
| |
| |-
| |
| |}
| |
|
| |
|
| == MIPAV DTI pipeline interface ==
| | TBD. |
|
| |
|
| ''Main article: [[DTI pipeline interface]].''
| | == Video tutorials == |
| | | TBD. |
| The MIPAV DTI Pipeline interface contains 6 tabs:
| |
| | |
| *In the [[DTI pipeline interface#ImportData |Import Data]] the user uploads DW and T2 image (optional), and the gradient information (optional). MIPAV could also read the gradient information from the image header. The Phillips Gradient Creator will then calculate the gradient table based on the information provided.
| |
| *In the [[DTI pipeline interface#PreProcessing | Pre-processing tab]], the user is asked to specify the parameters required for aligning B0 to T2, and then DWI to the rigidly aligned B0, and also parameters for motion correction and eddy current distortion correction.
| |
| * In the [[DTI pipeline interface #EPIDistortionCorrection | EPI Distortion Correction]], the user enters parameters needed for calculation of deformation vector fields for rigidly aligned B0 and T2 from the Pre-processing step.
| |
| * In the [[DTI pipeline interface #TensorEstimation |Tensor Estimation tab]], the user uploads the mask image, selects the tensor estimation algorithm and specifies the output options.
| |
| * In the [[DTI pipeline interface#TensorStatistics |Tensor Statistics]] the user can upload the tensor image and specify which kind of images (e.g. ADC, color map, Eigen value, Eigen vector, FA, RA, and Volume Ratio) he/she would like to have as an output.
| |
| * In the [[DTI pipeline interface#VisualizationTab |Visualization]], the user can upload images needed for a 3D visualization of fiber bundle tracts in the brain's white matter using the provided information. The user can then save fiber tracts information as [[Image formats descriptions#VtkXml |.vtk and .dat]] files.
| |
| | |
| == DTI Pipeline Tutorials == | |
| | |
| Refer to [[DTI Pipeline Tutorials]].
| |
|
| |
|
| TBD.
| | == See also:== |
|
| |
|
| == See also: ==
| | *[[DTI Pipeline| Main article: DTI Pipeline]] |
| | *[[DTI pipeline interface]] |
| *[[Understanding MIPAV capabilities]] | | *[[Understanding MIPAV capabilities]] |
| *[[Developing new tools using the API]] | | *[[Developing new tools using the API]] |